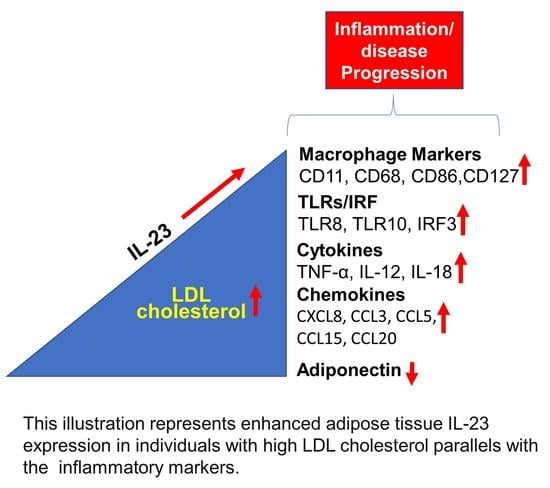
Increased Adipose Tissue Expression of IL-23 Associates with Inflammatory Markers in People with High LDL Cholesterol
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population and Anthropometric Measurements
2.2. Collection of Subcutaneous Adipose Tissue
2.3. Measurement of Biochemical Parameters
2.4. Real-Time qRT-PCR
2.5. Immunohistochemistry (IHC)
2.6. Statistical Analysis
3. Results
3.1. High IL-23 Expression Levels in Adipose Tissue of People with Increased Plasma Levels of LDL-C
3.2. Increased IL-23 Gene Expression in the Adipose Tissue Relates to Macrophage Markers
3.3. Increased Adipose IL-23 Gene Expression in People with High LDL-C Relates to TLR2-IRF3 Axis
3.4. Increased IL-23 Gene Expression in the Adipose Tissue Relates to Inflammatory Cytokines and Chemokines
3.5. Adipose IL-23 Gene Expression in People with H-LDL-C Associates Inversely with Plasma Adiponectin Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McKenzie, B.S.; Kastelein, R.A.; Cua, D.J. Understanding the il-23-il-17 immune pathway. Trends Immunol. 2006, 27, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Cauli, A.; Piga, M.; Floris, A.; Mathieu, A. Current perspective on the role of the interleukin-23/interleukin-17 axis in inflammation and disease (chronic arthritis and psoriasis). Immunotargets 2015, 4, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Gagro, A.; Servis, D.; Cepika, A.M.; Toellner, K.M.; Grafton, G.; Taylor, D.R.; Branica, S.; Gordon, J. Type i cytokine profiles of human naive and memory b lymphocytes: A potential for memory cells to impact polarization. Immunology 2006, 118, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Oppmann, B.; Lesley, R.; Blom, B.; Timans, J.C.; Xu, Y.; Hunte, B.; Vega, F.; Yu, N.; Wang, J.; Singh, K.; et al. Novel p19 protein engages il-12p40 to form a cytokine, il-23, with biological activities similar as well as distinct from il-12. Immunity 2000, 13, 715–725. [Google Scholar] [CrossRef]
- Teng, M.W.; Bowman, E.P.; McElwee, J.J.; Smyth, M.J.; Casanova, J.L.; Cooper, A.M.; Cua, D.J. Il-12 and il-23 cytokines: From discovery to targeted therapies for immune-mediated inflammatory diseases. Nat. Med. 2015, 21, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Allocca, M.; Furfaro, F.; Fiorino, G.; Gilardi, D.; D’Alessio, S.; Danese, S. Can il-23 be a good target for ulcerative colitis? Best Pract. Res. Clin. Gastroenterol. 2018, 32–33, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ji, S.; Cai, G.; Pan, Z.; Han, R.; Yuan, Y.; Xu, S.; Yang, J.; Hu, X.; Chen, M.; et al. H19 increases il-17a/il-23 releases via regulating vdr by interacting with mir675-5p/mir22-5p in ankylosing spondylitis. Mol. Ther. Nucleic Acids 2020, 19, 393–404. [Google Scholar] [CrossRef]
- Brentano, F.; Ospelt, C.; Stanczyk, J.; Gay, R.E.; Gay, S.; Kyburz, D. Abundant expression of the interleukin (il)23 subunit p19, but low levels of bioactive il23 in the rheumatoid synovium: Differential expression and toll-like receptor-(tlr) dependent regulation of the il23 subunits, p19 and p40, in rheumatoid arthritis. Ann. Rheum. Dis. 2009, 68, 143–150. [Google Scholar] [CrossRef]
- Liu, F.L.; Chen, C.H.; Chu, S.J.; Chen, J.H.; Lai, J.H.; Sytwu, H.K.; Chang, D.M. Interleukin (il)-23 p19 expression induced by il-1beta in human fibroblast-like synoviocytes with rheumatoid arthritis via active nuclear factor-kappab and ap-1 dependent pathway. Rheumatology 2007, 46, 1266–1273. [Google Scholar] [CrossRef]
- Yago, T.; Nanke, Y.; Kawamoto, M.; Furuya, T.; Kobashigawa, T.; Kamatani, N.; Kotake, S. Il-23 induces human osteoclastogenesis via il-17 in vitro, and anti-il-23 antibody attenuates collagen-induced arthritis in rats. Arthritis Res. Ther. 2007, 9, R96. [Google Scholar] [CrossRef] [Green Version]
- Moschen, A.R.; Tilg, H.; Raine, T. Il-12, il-23 and il-17 in ibd: Immunobiology and therapeutic targeting. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 185–196. [Google Scholar] [CrossRef]
- Sewell, G.W.; Kaser, A. Interleukin-23 in the pathogenesis of inflammatory bowel disease and implications for therapeutic intervention. J. Crohns Colitis 2022, 16, ii3–ii19. [Google Scholar] [CrossRef]
- Noviello, D.; Mager, R.; Roda, G.; Borroni, R.G.; Fiorino, G.; Vetrano, S. The il23-il17 immune axis in the treatment of ulcerative colitis: Successes, defeats, and ongoing challenges. Front. Immunol. 2021, 12, 611256. [Google Scholar] [CrossRef]
- Schmitt, H.; Neurath, M.F.; Atreya, R. Role of the il23/il17 pathway in crohn’s disease. Front. Immunol. 2021, 12, 622934. [Google Scholar] [CrossRef]
- Ziblat, A.; Nunez, S.Y.; Raffo Iraolagoitia, X.L.; Spallanzani, R.G.; Torres, N.I.; Sierra, J.M.; Secchiari, F.; Domaica, C.I.; Fuertes, M.B.; Zwirner, N.W. Interleukin (il)-23 stimulates ifn-gamma secretion by cd56(bright) natural killer cells and enhances il-18-driven dendritic cells activation. Front. Immunol. 2017, 8, 1959. [Google Scholar] [CrossRef]
- Aggarwal, S.; Ghilardi, N.; Xie, M.H.; de Sauvage, F.J.; Gurney, A.L. Interleukin-23 promotes a distinct cd4 t cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 2003, 278, 1910–1914. [Google Scholar] [CrossRef]
- Libby, P.; Ridker, P.M.; Hansson, G.K.; Leducq Transatlantic Network on Atherothrombosis. Inflammation in atherosclerosis: From pathophysiology to practice. J. Am. Coll. Cardiol. 2009, 54, 2129–2138. [Google Scholar] [CrossRef]
- Ridker, P.M. Residual inflammatory risk: Addressing the obverse side of the atherosclerosis prevention coin. Eur. Heart J. 2016, 37, 1720–1722. [Google Scholar] [CrossRef]
- Libby, P. Inflammation in atherosclerosis. Nature 2002, 420, 868–874. [Google Scholar] [CrossRef]
- Naruko, T.; Ueda, M.; Haze, K.; van der Wal, A.C.; van der Loos, C.M.; Itoh, A.; Komatsu, R.; Ikura, Y.; Ogami, M.; Shimada, Y.; et al. Neutrophil infiltration of culprit lesions in acute coronary syndromes. Circulation 2002, 106, 2894–2900. [Google Scholar] [CrossRef] [Green Version]
- Ben-Chetrit, E.; Levy, M. Colchicine: 1998 update. Semin. Arthritis Rheum. 1998, 28, 48–59. [Google Scholar] [CrossRef]
- Jukema, R.A.; Ahmed, T.A.N.; Tardif, J.C. Does low-density lipoprotein cholesterol induce inflammation? If so, does it matter? Current insights and future perspectives for novel therapies. BMC Med. 2019, 17, 197. [Google Scholar] [CrossRef]
- Abbas, A.; Gregersen, I.; Holm, S.; Daissormont, I.; Bjerkeli, V.; Krohg-Sorensen, K.; Skagen, K.R.; Dahl, T.B.; Russell, D.; Almas, T.; et al. Interleukin 23 levels are increased in carotid atherosclerosis: Possible role for the interleukin 23/interleukin 17 axis. Stroke 2015, 46, 793–799. [Google Scholar] [CrossRef]
- Manti, S.; Leonardi, S.; Panasiti, I.; Arrigo, T.; Salpietro, C.; Cuppari, C. Serum il-10, il-17 and il-23 levels as "bioumoral bridges" between dyslipidemia and atopy. Cytokine 2017, 99, 43–49. [Google Scholar] [CrossRef] [PubMed]
- National Cholesterol Education Program Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults. Third report of the national cholesterol education program (ncep) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel iii) final report. Circulation 2002, 106, 3143–3421. [Google Scholar] [CrossRef]
- Sindhu, S.; Thomas, R.; Shihab, P.; Sriraman, D.; Behbehani, K.; Ahmad, R. Obesity is a positive modulator of il-6r and il-6 expression in the subcutaneous adipose tissue: Significance for metabolic inflammation. PLoS ONE 2015, 10, e0133494. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Shihab, P.K.; Thomas, R.; Alghanim, M.; Hasan, A.; Sindhu, S.; Behbehani, K. Increased expression of the interleukin-1 receptor-associated kinase (irak)-1 is associated with adipose tissue inflammatory state in obesity. Diabetol. Metab. Syndr. 2015, 7, 71. [Google Scholar] [CrossRef]
- Knopfholz, J.; Disserol, C.C.; Pierin, A.J.; Schirr, F.L.; Streisky, L.; Takito, L.L.; Massucheto Ledesma, P.; Faria-Neto, J.R.; Olandoski, M.; da Cunha, C.L.; et al. Validation of the friedewald formula in patients with metabolic syndrome. Cholesterol 2014, 2014, 261878. [Google Scholar] [CrossRef]
- Ahmad, R.; Al-Mass, A.; Al-Ghawas, D.; Shareif, N.; Zghoul, N.; Melhem, M.; Hasan, A.; Al-Ghimlas, F.; Dermime, S.; Behbehani, K. Interaction of osteopontin with il-18 in obese individuals: Implications for insulin resistance. PLoS ONE 2013, 8, e63944. [Google Scholar] [CrossRef]
- Akhter, N.; Kochumon, S.; Hasan, A.; Wilson, A.; Nizam, R.; Al Madhoun, A.; Al-Rashed, F.; Arefanian, H.; Alzaid, F.; Sindhu, S.; et al. Ifn-gamma and lps induce synergistic expression of ccl2 in monocytic cells via h3k27 acetylation. J. Inflamm. Res. 2022, 15, 4291–4302. [Google Scholar] [CrossRef]
- Kochumon, S.; Jacob, T.; Koshy, M.; Al-Rashed, F.; Sindhu, S.; Al-Ozairi, E.; Al-Mulla, F.; Rosen, E.D.; Ahmad, R. Palmitate potentiates lipopolysaccharide-induced il-6 production via coordinated acetylation of h3k9/h3k18, p300, and rna polymerase ii. J. Immunol. 2022, 209, 731–741. [Google Scholar]
- Russo, L.; Lumeng, C.N. Properties and functions of adipose tissue macrophages in obesity. Immunology 2018, 155, 407–417. [Google Scholar] [CrossRef]
- Ahmad, R.; Al-Mass, A.; Atizado, V.; Al-Hubail, A.; Al-Ghimlas, F.; Al-Arouj, M.; Bennakhi, A.; Dermime, S.; Behbehani, K. Elevated expression of the toll like receptors 2 and 4 in obese individuals: Its significance for obesity-induced inflammation. J. Inflamm. 2012, 9, 48. [Google Scholar] [CrossRef]
- Sindhu, S.; Akhter, N.; Kochumon, S.; Thomas, R.; Wilson, A.; Shenouda, S.; Tuomilehto, J.; Ahmad, R. Increased expression of the innate immune receptor tlr10 in obesity and type-2 diabetes: Association with ros-mediated oxidative stress. Cell Physiol. Biochem. 2018, 45, 572–590. [Google Scholar] [CrossRef]
- Izadi, V.; Farabad, E.; Azadbakht, L. Epidemiologic evidence on serum adiponectin level and lipid profile. Int. J. Prev. Med. 2013, 4, 133–140. [Google Scholar]
- Ye, J.; Wang, Y.; Wang, Z.; Liu, L.; Yang, Z.; Wang, M.; Xu, Y.; Ye, D.; Zhang, J.; Lin, Y.; et al. Roles and mechanisms of interleukin-12 family members in cardiovascular diseases: Opportunities and challenges. Front. Pharmacol. 2020, 11, 129. [Google Scholar] [CrossRef]
- Mortensen, M.B.; Nordestgaard, B.G. Elevated ldl cholesterol and increased risk of myocardial infarction and atherosclerotic cardiovascular disease in individuals aged 70-100 years: A contemporary primary prevention cohort. Lancet 2020, 396, 1644–1652. [Google Scholar] [CrossRef]
- Fatima, N.; Faisal, S.M.; Zubair, S.; Siddiqui, S.S.; Moin, S.; Owais, M. Emerging role of interleukins il-23/il-17 axis and biochemical markers in the pathogenesis of type 2 diabetes: Association with age and gender in human subjects. Int. J. Biol. Macromol. 2017, 105, 1279–1288. [Google Scholar] [CrossRef]
- Ye, J.; Wang, Y.; Wang, Z.; Liu, L.; Yang, Z.; Wang, M.; Xu, Y.; Ye, D.; Zhang, J.; Zhou, Q.; et al. The expression of il-12 family members in patients with hypertension and its association with the occurrence of carotid atherosclerosis. Mediat. Inflamm. 2020, 2020, 2369279. [Google Scholar] [CrossRef]
- Libby, P.; Buring, J.E.; Badimon, L.; Hansson, G.K.; Deanfield, J.; Bittencourt, M.S.; Tokgozoglu, L.; Lewis, E.F. Atherosclerosis. Nat. Rev. Dis. Primers 2019, 5, 56. [Google Scholar] [CrossRef]
- Bobryshev, Y.V.; Ivanova, E.A.; Chistiakov, D.A.; Nikiforov, N.G.; Orekhov, A.N. Macrophages and their role in atherosclerosis: Pathophysiology and transcriptome analysis. BioMed Res. Int. 2016, 2016, 9582430. [Google Scholar] [CrossRef]
- Mortensen, M.B.; Cainzos-Achirica, M.; Steffensen, F.H.; Botker, H.E.; Jensen, J.M.; Sand, N.P.R.; Maeng, M.; Bruun, J.M.; Blaha, M.J.; Sorensen, H.T.; et al. Association of coronary plaque with low-density lipoprotein cholesterol levels and rates of cardiovascular disease events among symptomatic adults. JAMA Netw. Open 2022, 5, e2148139. [Google Scholar] [CrossRef]
- Bednarska, K.; Kielbik, M.; Sulowska, Z.; Dziadek, J.; Klink, M. Cholesterol oxidase binds tlr2 and modulates functional responses of human macrophages. Mediat. Inflamm. 2014, 2014, 498395. [Google Scholar] [CrossRef]
- Schoneveld, A.H.; Oude Nijhuis, M.M.; van Middelaar, B.; Laman, J.D.; de Kleijn, D.P.; Pasterkamp, G. Toll-like receptor 2 stimulation induces intimal hyperplasia and atherosclerotic lesion development. Cardiovasc. Res. 2005, 66, 162–169. [Google Scholar] [CrossRef]
- Mullick, A.E.; Tobias, P.S.; Curtiss, L.K. Modulation of atherosclerosis in mice by toll-like receptor 2. J. Clin. Investig. 2005, 115, 3149–3156. [Google Scholar] [CrossRef]
- Ahmad, R.; Al-Roub, A.; Kochumon, S.; Akther, N.; Thomas, R.; Kumari, M.; Koshy, M.S.; Tiss, A.; Hannun, Y.A.; Tuomilehto, J.; et al. The synergy between palmitate and tnf-alpha for ccl2 production is dependent on the trif/irf3 pathway: Implications for metabolic inflammation. J. Immunol. 2018, 200, 3599–3611. [Google Scholar] [CrossRef]
- Patel, S.J.; Liu, N.; Piaker, S.; Gulko, A.; Andrade, M.L.; Heyward, F.D.; Sermersheim, T.; Edinger, N.; Srinivasan, H.; Emont, M.P.; et al. Hepatic irf3 fuels dysglycemia in obesity through direct regulation of ppp2r1b. Sci. Transl. Med. 2022, 14, eabh3831. [Google Scholar] [CrossRef]
- Kumari, M.; Wang, X.; Lantier, L.; Lyubetskaya, A.; Eguchi, J.; Kang, S.; Tenen, D.; Roh, H.C.; Kong, X.; Kazak, L.; et al. Irf3 promotes adipose inflammation and insulin resistance and represses browning. J. Clin. Investig. 2016, 126, 2839–2854. [Google Scholar] [CrossRef]
- Kochumon, S.; Arefanian, H.; Azim, R.; Shenouda, S.; Jacob, T.; Abu Khalaf, N.; Al-Rashed, F.; Hasan, A.; Sindhu, S.; Al-Mulla, F.; et al. Stearic acid and tnf-alpha co-operatively potentiate mip-1alpha production in monocytic cells via myd88 independent tlr4/tbk/irf3 signaling pathway. Biomedicines 2020, 8, 403. [Google Scholar] [CrossRef]
- Eguchi, J.; Kong, X.; Tenta, M.; Wang, X.; Kang, S.; Rosen, E.D. Interferon regulatory factor 4 regulates obesity-induced inflammation through regulation of adipose tissue macrophage polarization. Diabetes 2013, 62, 3394–3403. [Google Scholar] [CrossRef]
- Wieser, V.; Moschen, A.R.; Tilg, H. Inflammation, cytokines and insulin resistance: A clinical perspective. Arch. Immunol. Ther. Exp. 2013, 61, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Boesten, L.S.; Zadelaar, A.S.; van Nieuwkoop, A.; Gijbels, M.J.; de Winther, M.P.; Havekes, L.M.; van Vlijmen, B.J. Tumor necrosis factor-alpha promotes atherosclerotic lesion progression in apoe*3-leiden transgenic mice. Cardiovasc. Res. 2005, 66, 179–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Sun, C.; Gerdes, N.; Liu, C.; Liao, M.; Liu, J.; Shi, M.A.; He, A.; Zhou, Y.; Sukhova, G.K.; et al. Interleukin 18 function in atherosclerosis is mediated by the interleukin 18 receptor and the na-cl co-transporter. Nat. Med. 2015, 21, 820–826. [Google Scholar] [CrossRef] [PubMed]
- De Groote, D.; Zangerle, P.F.; Gevaert, Y.; Fassotte, M.F.; Beguin, Y.; Noizat-Pirenne, F.; Pirenne, J.; Gathy, R.; Lopez, M.; Dehart, I.; et al. Direct stimulation of cytokines (il-1 beta, tnf-alpha, il-6, il-2, ifn-gamma and gm-csf) in whole blood. I. Comparison with isolated pbmc stimulation. Cytokine 1992, 4, 239–248. [Google Scholar] [CrossRef]
- Bardina, S.V.; Michlmayr, D.; Hoffman, K.W.; Obara, C.J.; Sum, J.; Charo, I.F.; Lu, W.; Pletnev, A.G.; Lim, J.K. Differential roles of chemokines ccl2 and ccl7 in monocytosis and leukocyte migration during west nile virus infection. J. Immunol. 2015, 195, 4306–4318. [Google Scholar] [CrossRef] [PubMed]
- Koenen, R.R.; Weber, C. Chemokines: Established and novel targets in atherosclerosis. EMBO Mol. Med. 2011, 3, 713–725. [Google Scholar] [CrossRef]
- de Jager, S.C.; Bongaerts, B.W.; Weber, M.; Kraaijeveld, A.O.; Rousch, M.; Dimmeler, S.; van Dieijen-Visser, M.P.; Cleutjens, K.B.; Nelemans, P.J.; van Berkel, T.J.; et al. Chemokines ccl3/mip1alpha, ccl5/rantes and ccl18/parc are independent risk predictors of short-term mortality in patients with acute coronary syndromes. PLoS ONE 2012, 7, e45804. [Google Scholar] [CrossRef]
- Gibaldi, D.; Vilar-Pereira, G.; Pereira, I.R.; Silva, A.A.; Barrios, L.C.; Ramos, I.P.; Mata Dos Santos, H.A.; Gazzinelli, R.; Lannes-Vieira, J. Ccl3/macrophage inflammatory protein-1alpha is dually involved in parasite persistence and induction of a tnf- and ifngamma-enriched inflammatory milieu in trypanosoma cruzi-induced chronic cardiomyopathy. Front. Immunol. 2020, 11, 306. [Google Scholar] [CrossRef]
- de Jager, S.C.; Bot, I.; Kraaijeveld, A.O.; Korporaal, S.J.; Bot, M.; van Santbrink, P.J.; van Berkel, T.J.; Kuiper, J.; Biessen, E.A. Leukocyte-specific ccl3 deficiency inhibits atherosclerotic lesion development by affecting neutrophil accumulation. Arterioscler. Thromb. Vasc. Biol. 2013, 33, e75–e83. [Google Scholar] [CrossRef]
- Wang, C.; Zou, Y.; Pan, C.; Shao, L.; Ding, Z.; Zhang, Y.; Ye, J.; Li, P.; Ren, Y.; Zhu, C. Prognostic significance of chemokines ccl11 and ccl5 modulated by low-density lipoprotein cholesterol in colon cancer patients with normal body mass index. Ann. Transl. Med. 2021, 9, 202. [Google Scholar] [CrossRef]
- Orekhov, A.N.; Nikiforov, N.G.; Elizova, N.V.; Korobov, G.A.; Aladinskaya, A.V.; Sobenin, I.A.; Bobryshev, Y.V. Tumor necrosis factor-alpha and c-c motif chemokine ligand 18 associate with atherosclerotic lipid accumulation in situ and in vitro. Curr. Pharm. Des. 2018, 24, 2883–2889. [Google Scholar] [CrossRef]





| Low LDL-c (n = 24) | High LDL-c (n = 36) | p-Value | |
|---|---|---|---|
| Median (IQR) | Median (IQR) | ||
| Age | 44.5 (36.00–53.50) | 43 (35.25–50.5) | 0.561 |
| Weight (kg) | 88.65 (68.25–99.20) | 83.2 (73.18–92.38) | 0.248 |
| Height (m) | 1.65 (1.57–1.73) | 1.67 (1.56–1.74) | 0.493 |
| Waist (cm) | 104.0 (90.50–114.30) | 96.5 (87.25–108.0) | 0.181 |
| Hip (cm) | 111.0 (103.30–125.00) | 108.50 (99.50–116.00) | 0.187 |
| WHR | 0.89.0 (0.81–1.02) | 0.89.0 (0.79–0.99) | 0.77 |
| BMI (kg/m2) | 31.62 (27.39–37.35) | 29.86 (27.0–34.22) | 0.151 |
| PBF (%) | 38.10 (32.85–44.05) | 36.20 (28.80–38.70) | 0.052 |
| GLUC (mmol/l) | 5.05 (4.75–5.42) | 5.10 (4.80–5.74) | 0.536 |
| TGL (mmol/l) | 0.95 (0.58–1.85) | 0.97 (0.68–1.50) | 0.689 |
| Chol (mmol/l) | 4.22 (3.70–4.68) | 5.54 (5.00–6.14) | <0.0001 |
| HDL-c (mmol/l) | 1.16 (0.99–1.38) | 1.25 (1.05–1.49) | 0.268 |
| LDL-c (mmol/l) | 2.55 (2.30–2.70) | 3.70 (3.23–4.00) | <0.0001 |
| HbA1C (%) | 5.35 (5.16–5.90) | 5.70 (5.50–5.90) | 0.03 |
| Insulin (mU/L) | 6.11 (5.50–10.44) | 5.33 (4.89–7.32) | 0.027 |
| HOMA-IR | 1.54 (1.18–2.58) | 1.32 (1.11–2.17) | 0.37 |
| Gene | Assay ID | Gene | Assay ID | Gene | Assay ID |
|---|---|---|---|---|---|
| IL-1β | Hs01555410_m1 | CCL8 | Hs04187715_m1 | CD141 | Hs00264920_s1 |
| IL-2 | Hs00174114_m1 | CCL-11 | Hs00237013_m1 | CD163 | Hs00174705_m1 |
| IL-5 | Hs01548712_g1 | CCL-15 | Hs00361122_m1 | CD302 | Hs00994886_m1 |
| IL-6 | Hs00985639_m1 | CCL18 | Hs00268113_m1 | TLR2 | Hs01872448_s1 |
| IL-8 | Hs00174103_m1 | CCL-19 | Hs00171149_m1 | TLR3 | Hs01551078_m1 |
| IL-10 | Hs00961622_m1 | CCL-20 | Hs01011368_m1 | TLR4 | Hs00152939_m1 |
| IL-12A | Hs01073447_m1 | CXCL9 | Hs00171065_m1 | TLR7 | Hs01933259_s1 |
| IL-13 | Hs00174379_m1 | CXCL10 | Hs01124251_g1 | TLR8 | Hs00152972_m1 |
| IL-18 | Hs01038788_m1 | CXCL-11 | Hs04187682_g1 | TLR9 | Hs00370913_s1 |
| IL-33 | Hs00369211_m1 | IL-1RL1 | Hs00545033_m1 | TLR10 | Hs01935337_s1 |
| TNF-α | Hs01113624_g1 | IL-2RA | Hs00907779_m1 | IRF3 | Hs01547283_m1 |
| TGF-β | Hs00820148_g1 | CCR1 | Hs00928897_s1 | IRF4 | Hs01056533_m1 |
| IFNB1 | Hs01077958_s1 | CCR2 | Hs00704702_s1 | IRF5 | Hs00158114_m1 |
| Dectin | Hs01902549_s1 | CCR5 | Hs99999149_s1 | MyD88 | Hs01573837_g1 |
| SRA1 | Hs00398296_g1 | CD11c | Hs00174217_m1 | IRAK1 | Hs01018347_m1 |
| CCL2 | Hs00234140_m1 | CD16 | Hs04334165_m1 | TRAF6 | Hs00371512_g1 |
| CCL3 | Hs04194942_s1 | CD68 | Hs02836816_g1 | NFKB | Hs00765730_m1 |
| CCL5 | Hs00982282_m1 | CD86 | Hs01567026_m1 | GAPDH | Hs03929097_g1 |
| CCL-7 | Hs00171147_m1 | IL7R | Hs00902334_m1 |
| Macrophage Markers | L-LDL-c | H-LDL-c | ||
|---|---|---|---|---|
| IL-23 | r | p | r | p |
| ITGAX (CD11c) | 0.2617 | 0.2651 | 0.5625 | 0.0004 * |
| CD16 | 0.4831 | 0.0265* | 0.5944 | 0.0001 * |
| CD68 | 0.2429 | 0.2886 | 0.397 | 0.024 * |
| CD86 | 0.3158 | 0.175 | 0.3708 | 0.0283 * |
| CD127/IL7R | 0.04025 | 0.874 | 0.5739 | 0.0004 * |
| CD141 | −0.06028 | 0.7847 | 0.3252 | 0.0648 |
| CD163 | 0.5325 | 0.0107 * | 0.2899 | 0.0864 |
| CD302 | 0.2226 | 0.2958 | 0.1278 | 0.4576 |
| L-LDL-c | H-LDL-c | |||
|---|---|---|---|---|
| IL-23 | r | p | r | p |
| TLR2 | 0.415 | 0.069 | 0.485 | 0.008 * |
| TLR3 | 0.116 | 0.627 | 0.062 | 0.736 |
| TLR4 | 0.015 | 0.951 | 0.017 | 0.928 |
| TLR7 | 0.339 | 0.114 | 0.209 | 0.221 |
| TLR9 | 0.150 | 0.483 | 0.024 | 0.891 |
| IRF3 | −0.132 | 0.612 | 0.464 | 0.010 * |
| IRF4 | 0.330 | 0.144 | 0.129 | 0.490 |
| IRF5 | 0.253 | 0.244 | 0.186 | 0.300 |
| MyD88 | 0.244 | 0.262 | 0.182 | 0.295 |
| IRAK1 | −0.110 | 0.618 | 0.201 | 0.261 |
| TRAF6 | 0.390 | 0.059 | −0.025 | 0.885 |
| Cytokines/Chemokines | L-LDL-c | H-LDL-c | ||
|---|---|---|---|---|
| IL-23 | r | p | r | p |
| IL-1β | −0.250 | 0.349 | 0.178 | 0.364 |
| IL-2 | 0.504 | 0.017 * | 0.200 | 0.242 |
| IL-5 | −0.347 | 0.105 | −0.220 | 0.235 |
| IL-6 | 0.060 | 0.789 | −0.070 | 0.708 |
| IL-8 | 0.079 | 0.733 | 0.314 | 0.091 |
| IL-10 | 0.209 | 0.363 | 0.097 | 0.581 |
| IL-12A | −0.081 | 0.785 | 0.516 | 0.004 * |
| IL-13 | 0.204 | 0.375 | 0.030 | 0.872 |
| IL-18 | 0.122 | 0.598 | 0.350 | 0.043 * |
| TNF-α | 0.318 | 0.160 | 0.375 | 0.032 * |
| TGF-β | 0.122 | 0.609 | 0.396 | 0.018 * |
| IFNB1 | 0.172 | 0.421 | 0.398 | 0.024 * |
| CCL2 | 0.214 | 0.316 | 0.130 | 0.471 |
| CCL3 | 0.392 | 0.079 | 0.426 | 0.017 * |
| CCL5 | 0.323 | 0.191 | 0.632 | 0.0001 * |
| CCL-7 | 0.189 | 0.388 | 0.173 | 0.336 |
| CCL-11 | 0.096 | 0.679 | −0.066 | 0.709 |
| CCL-15 | 0.163 | 0.457 | 0.370 | 0.026 * |
| CCL-19 | 0.023 | 0.920 | 0.078 | 0.656 |
| CCL-20 | 0.171 | 0.436 | 0.336 | 0.049 * |
| CXCL9 | 0.056 | 0.799 | 0.002 | 0.991 |
| CXCL10 | 0.162 | 0.460 | 0.205 | 0.245 |
| Metabolic Markers | L-LDL | H-LDL | ||
|---|---|---|---|---|
| r | p | r | p | |
| BMI | −0.1696 | 0.4283 | 0.04376 | 0.7999 |
| PBF | −0.05974 | 0.797 | −0.1982 | 0.2851 |
| Waist | −0.1702 | 0.4606 | 0.1198 | 0.5138 |
| Hip | −0.1419 | 0.5396 | 0.1568 | 0.3916 |
| Chol | −0.00914 | 0.9662 | 0.09003 | 0.6016 |
| HDL | −0.06958 | 0.7466 | 0.3282 | 0.0507 |
| LDL | 0.07818 | 0.7165 | −0.171 | 0.3188 |
| TGL | −0.1591 | 0.4577 | 0.1827 | 0.2863 |
| GLU | 0.1319 | 0.5391 | −0.05659 | 0.7431 |
| HBA1C | 0.006998 | 0.9741 | 0.1795 | 0.3021 |
| WBC | −0.02482 | 0.9083 | −0.2334 | 0.184 |
| Adiponectin | −0.264 | 0.34 | −0.437 | 0.0372 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kochumon, S.; Hasan, A.; Al-Rashed, F.; Sindhu, S.; Thomas, R.; Jacob, T.; Al-Sayyar, A.; Arefanian, H.; Al Madhoun, A.; Al-Ozairi, E.; et al. Increased Adipose Tissue Expression of IL-23 Associates with Inflammatory Markers in People with High LDL Cholesterol. Cells 2022, 11, 3072. https://doi.org/10.3390/cells11193072
Kochumon S, Hasan A, Al-Rashed F, Sindhu S, Thomas R, Jacob T, Al-Sayyar A, Arefanian H, Al Madhoun A, Al-Ozairi E, et al. Increased Adipose Tissue Expression of IL-23 Associates with Inflammatory Markers in People with High LDL Cholesterol. Cells. 2022; 11(19):3072. https://doi.org/10.3390/cells11193072
Chicago/Turabian StyleKochumon, Shihab, Amal Hasan, Fatema Al-Rashed, Sardar Sindhu, Reeby Thomas, Texy Jacob, Amnah Al-Sayyar, Hossein Arefanian, Ashraf Al Madhoun, Ebaa Al-Ozairi, and et al. 2022. "Increased Adipose Tissue Expression of IL-23 Associates with Inflammatory Markers in People with High LDL Cholesterol" Cells 11, no. 19: 3072. https://doi.org/10.3390/cells11193072