The Olfactory Receptor Gene Product, OR5H2, Modulates Endometrial Cancer Cells Proliferation via Interaction with the IGF1 Signaling Pathway
Abstract
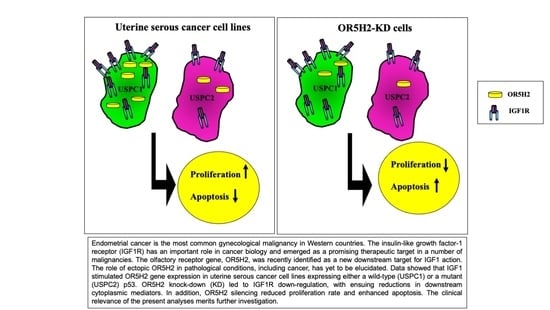
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Treatments
2.2. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.3. Western Blot Analysis
2.4. Small-Interfering RNA (siRNA) OR5H2 Knockdown
2.5. Proliferation Assays
2.6. Cell Cycle Analysis
2.7. Co-Immunoprecipitation (Co-IP) Assays
2.8. Animal Studies
2.9. Statistical Analysis
3. Results
3.1. Identification of OR5H2 as a Target for IGF1 Action
3.2. Regulation of OR5H2 Gene Expression by IGF1 and Insulin in Endometrial Cancer Cells
3.3. Effect of OR5H2 Knockdown on the IGF1R Signaling Pathway
3.4. Co-Immunoprecipitation of IGF1R and OR5H2
3.5. Effect of OR5H2 Knockdown on Endometrial Cell Proliferation
3.6. Effect of OR5H2 Knockdown on Cell Cycle Dynamics
3.7. Animal Studies
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yakar, S.; Adamo, M.L. Insulin-like growth factor 1 physiology: Lessons from mouse models. Endocrinol. Metab. Clin. N. Am. 2012, 41, 231–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeRoith, D. Clinical relevance of systemic and local IGF-I: Lessons from animal models. Pediatr. Endocrinol. Rev. 2008, 5, 739–743. [Google Scholar] [PubMed]
- Holly, J.M.; Perks, C.M. Insulin-like growth factor physiology: What we have learned from human studies. Endocrinol. Metab. Clin. N. Am. 2012, 41, 249–263. [Google Scholar] [CrossRef]
- Chhabra, Y.; Lee, C.M.M.; Müller, A.F.; Brooks, A.J. GHR signalling: Receptor activation and degradation mechanisms. Mol. Cell. Endocrinol. 2021, 520, 111075. [Google Scholar] [CrossRef]
- Waters, M.J. The growth hormone receptor. Growth Horm. IGF Res. 2016, 28, 6–10. [Google Scholar] [CrossRef] [Green Version]
- Werner, H.; Sarfstein, R. Transcriptional and epigenetic control of IGF1R gene expression: Implications in metabolism and cancer. Growth Horm. IGF Res. 2014, 24, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Girnita, L.; Worrall, C.; Takahashi, S.; Seregard, S.; Girnita, A. Something old, something new and something borrowed: Emerging paradigm of insulin-like growth factor type 1 receptor (IGF-1R) signaling regulation. Cell. Mol. Life Sci. 2014, 71, 2403–2427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baserga, R.; Peruzzi, F.; Reiss, K. The IGF-1 receptor in cancer biology. Int. J. Cancer 2003, 107, 873–877. [Google Scholar] [CrossRef] [PubMed]
- Klammt, J.; Pfaffle, R.; Werner, H.; Kiess, W. IGF signaling defects as causes of growth failure and IUGR. Trends Endocrinol. Metab. 2008, 19, 197–205. [Google Scholar] [CrossRef]
- Domené, S.; Domené, H.M. Genetic mutations in the GH/IGF axis. Pediatr. Endocrinol. Rev. 2018, 16, 39–62. [Google Scholar] [PubMed]
- Laron, Z. Extensive personal experience. Laron syndrome (primary growth hormone resistance or insensitivity): The personal experience 1958–2003. J. Clin. Endocrinol. Metab. 2004, 89, 1031–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laron, Z.; Pertzelan, A.; Mannheimer, S. Genetic pituitary dwarfism with high serum concentration of growth hormone—A new inborn error of metabolism? Isr. J. Med. Sci. 1966, 2, 152–155. [Google Scholar] [PubMed]
- Amselem, S.; Duquesnoy, P.; Attree, O.; Novelli, G.; Bousnina, S.; Postel-Vinay, M.C.; Goossens, M. Laron dwarfism and mutations of the growth hormone-receptor gene. N. Engl. J. Med. 1989, 321, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Laron, Z.; Kopchik, J.J. Laron Syndrome—From Man to Mouse; Springer: Berlin, Germany, 2011. [Google Scholar]
- Shevah, O.; Laron, Z. Patients with congenital deficiency of IGF-I seem protected from the development of malignancies: A preliminary report. Growth Horm. IGF Res. 2007, 17, 54–57. [Google Scholar] [CrossRef]
- Steuerman, R.; Shevah, O.; Laron, Z. Congenital IGF1 deficiency tends to confer protection against post-natal development of malignancies. Eur. J. Endocrinol. 2011, 164, 485–489. [Google Scholar] [CrossRef] [Green Version]
- Guevara-Aguirre, J.; Balasubramanian, P.; Guevara-Aguirre, M.; Wei, M.; Madia, F.; Cheng, C.W.; Hwang, D.; Martin-Montalvo, A.; Saavedra, J.; Ingles, S.; et al. Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci. Transl. Med. 2011, 3, 70ra13. [Google Scholar] [CrossRef] [Green Version]
- Werner, H.; Sarfstein, R.; Nagaraj, K.; Laron, Z. Laron syndrome research paves the way for new insights in oncological investigation. Cells 2020, 9, 2446. [Google Scholar] [CrossRef] [PubMed]
- Lapkina-Gendler, L.; Rotem, I.; Pasmanik-Chor, M.; Gurwitz, D.; Sarfstein, R.; Laron, Z.; Werner, H. Identification of signaling pathways associated with cancer protection in Laron syndrome. Endocr. Relat. Cancer 2016, 23, 399–410. [Google Scholar] [CrossRef] [Green Version]
- Werner, H.; Lapkina-Gendler, L.; Achlaug, L.; Nagaraj, K.; Somri, L.; Yaron-Saminsky, D.; Pasmanik-Chor, M.; Sarfstein, R.; Laron, Z.; Yakar, S. Genome-wide profiling of Laron syndrome patients identifies novel cancer protection pathways. Cells 2019, 8, 596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antunes, G.; Simoes de Souza, F.M. Olfactory receptor signaling. Methods Cell Biol. 2016, 132, 127–145. [Google Scholar] [PubMed]
- Massberg, D.; Hatt, H. Human olfactory receptors: Novel cellular functions outside of the nose. Physiol. Rev. 2018, 98, 1739–1763. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, H.; Fu, N.; Chen, L. The diversified function and potential therapy of ectopic olfactory receptors in non-olfactory tissues. J. Cell. Physiol. 2018, 233, 2104–2115. [Google Scholar] [CrossRef]
- Weng, J.; Wang, J.; Hu, X.; Wang, F.; Ittmann, M.; Liu, M. PSGR2, a novel G-protein coupled receptor, is overexpressed in human prostate cancer. Int. J. Cancer 2006, 118, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.L.; Sun, C.; Petrovics, G.; Makarem, M.; Furusato, B.; Zhang, W.; Sesterhenn, I.A.; McLeod, D.G.; Sun, L.; Moul, J.W.; et al. Quantitative expression profile of PSGR in prostate cancer. Prostate Cancer Prostatic Dis. 2006, 9, 56–61. [Google Scholar] [CrossRef] [Green Version]
- Morita, R.; Hirohashi, Y.; Torigoe, T.; Ito-Inoda, S.; Takahashi, A.; Mariya, T.; Asanuma, H.; Tamura, Y.; Tsukahara, T.; Kanaseki, T.; et al. Olfactory receptor family 7 subfamily C member 1 is a novel marker of colon cancer-initiating cells and is a potent target of immunotherapy. Clin. Cancer Res. 2016, 22, 3298–3309. [Google Scholar] [CrossRef] [Green Version]
- Giandomenico, V.; Cui, T.; Grimelius, L.; Öberg, K.; Pelosi, G.; Tsolakis, A.V. Olfactory receptor 51E1 as a novel target for diagnosis in somatostatin receptor-negative lung carcinoids. J. Mol. Endocrinol. 2013, 51, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Cui, T.; Tsolakis, A.V.; Li, S.C.; Cunningham, J.L.; Lind, T.; Öberg, K.; Giandomenico, V. Olfactory receptor 51E1 protein as a potential novel tissue biomarker for small intestine neuroendocrine carcinomas. Eur. J. Endocrinol. 2013, 168, 253–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masjedi, S.; Zwiebel, L.J.; Giorgio, T.D. Olfactory receptor gene abundance in invasive breast carcinoma. Sci. Rep. 2019, 9, 13736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goff, B.A.; Kato, D.; Schmidt, R.A.; Ek, M.; Ferry, J.A.; Muntz, H.G.; Cain, J.M.; Tamimi, H.K.; Figge, D.C.; Greer, B.E. Uterine papillary serous carcinoma: Patterns of metastatic spread. Gynecol. Oncol. 1994, 54, 264–268. [Google Scholar] [CrossRef]
- Hamilton, C.A.; Cheung, M.K.; Osann, K.; Chen, L.; Teng, N.N.; Longacre, T.A.; Powell, M.A.; Hendrickson, M.R.; Kapp, D.S.; Chan, J.K. Uterine papillary serous and clear cell carcinomas predict for poorer survival compared to grade 3 endometrioid corpus cancers. Br. J. Cancer 2006, 94, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Liu, Y.; Yi, X.; Miron, A.; Crum, C.; Kong, B.; Zheng, W. Endometrial glandular dysplasia with frequent p53 gene mutation: A genetic evidence supporting its precancer nature for endometrial serous carcinoma. Clin. Cancer Res. 2008, 14, 2263–2269. [Google Scholar] [CrossRef] [Green Version]
- Bruchim, I.; Werner, H. Targeting IGF-1 signaling pathways in gynecologic malignancies. Expert Opin. Ther. Targets 2013, 17, 307–320. [Google Scholar] [CrossRef]
- Irwin, J.C.; de las Fuentes, L.; Dsupin, B.A.; Giudice, L.C. Insulin-like growth factor regulation of human endometrial stromal cell function: Coordinate effects on insulin-like growth factor binding protein-1, cell proliferation and prolactin secretion. Regul. Pept. 1993, 48, 165–177. [Google Scholar] [CrossRef]
- Bruchim, I.; Sarfstein, R.; Werner, H. The IGF hormonal network in endometrial cancer: Functions, regulation, and targeting approaches. Front. Endocrinol. 2014, 5, 76. [Google Scholar] [CrossRef] [Green Version]
- Attias-Geva, Z.; Bentov, I.; Kidron, D.; Amichay, K.; Sarfstein, R.; Fishman, A.; Bruchim, I.; Werner, H. p53 regulates insulin-like growth factor-I receptor gene expression in uterine serous carcinoma and predicts responsiveness to an insulin-like growth factor-I receptor-directed targeted therapy. Eur. J. Cancer 2012, 48, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Santin, A.D.; Bellone, S.; Godken, M.; Palmieri, M.; Dunn, D.; Agha, J.; Roman, J.J.; Hutchins, L.; Pecorelli, S.; O’Brien, T.; et al. Overexpression of HER-2/Neu in uterine serous papillary cancer. Clin. Cancer Res. 2002, 8, 1271–1279. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Werner, H.; Laron, Z. Role of the GH-IGF1 system in progression of cancer. Mol. Cell. Endocrinol. 2020, 518, 111003. [Google Scholar] [CrossRef]
- Renehan, A.G.; Zwahlen, M.; Minder, C.; O’Dwyer, S.T.; Shalet, S.M.; Egger, M. Insulin-like growth factor-I, IGF binding protein-3, and cancer risk: Systematic review and meta-regression analysis. Lancet 2004, 363, 1346–1353. [Google Scholar] [CrossRef]
- Chan, J.M.; Stampfer, M.J.; Giovannucci, E.; Gann, P.H.; Ma, J.; Wilkinson, P.; Hennekens, C.H.; Pollak, M. Plasma insulin-like growth factor-I and prostate cancer risk: A prospective study. Science 1998, 279, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Hankinson, S.E.; Willett, W.C.; Colditz, G.A.; Hunter, D.J.; Michaud, D.S.; Deroo, B.; Rosner, B.; Speizer, F.E.; Pollak, M. Circulating concentrations of insulin-like growth factor-I and risk of breast cancer. Lancet 1998, 351, 1393–1396. [Google Scholar] [CrossRef]
- Hirano, S.; Ito, N.; Takahashi, S.; Tamaya, T. Clinical implications of insulin-like growth factors through the presence of their binding proteins and receptors expressed in gynecological cancers. Eur. J. Gynaecol. Oncol. 2004, 25, 187–191. [Google Scholar]
- Sinai-Livne, T.; Pasmanik-Chor, M.; Cohen, Z.; Tsarfaty, I.; Werner, H.; Berger, R. Proteomic analysis of combined IGF1 receptor targeted therapy and chemotherapy identifies signatures associated with survival in breast cancer patients. Oncotarget 2020, 11, 1515–1530. [Google Scholar] [CrossRef] [PubMed]
- Sanz, G.; Leray, I.; Muscat, A.; Acquistapace, A.; Cui, T.; Rivière, J.; Vincent-Naulleau, S.; Giandomenico, V.; Mir, L.M. Gallein, a Gβγ subunit signalling inhibitor, inhibits metastatic spread of tumour cells expressing OR51E2 and exposed to its odorant ligand. BMC Res. Notes 2017, 10, 541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pronin, A.; Slepak, V. Ectopically expressed olfactory receptors OR51E1 and OR51E2 suppress proliferation and promote cell death in a prostate cancer cell line. J. Biol. Chem. 2021, 296, 100475. [Google Scholar] [CrossRef] [PubMed]
- Girnita, A.; Girnita, L.; del Prete, F.; Bartolazzi, A.; Larsson, O.; Axelson, M. Cyclolignans as inhibitors of the insulin-like growth factor-1 receptor and malignant cell growth. Cancer Res. 2004, 64, 236–242. [Google Scholar] [CrossRef] [Green Version]
- Macaulay, V.M.; Middleton, M.R.; Eckhardt, S.G.; Rudin, C.M.; Juergens, R.A.; Gedrich, R.; Gogov, S.; McCarthy, S.; Poondru, S.; Stephens, A.W.; et al. Phase I dose-escalation study of Linsitinib (OSI-906) and Erlotinib in patients with advanced solid tumors. Clin. Cancer Res. 2016, 22, 2897–2907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scotlandi, K.; Manara, M.C.; Nicoletti, G.; Lollini, P.L.; Lukas, S.; Benini, S.; Croci, S.; Perdichizzi, S.; Zambelli, D.; Serra, M.; et al. Antitumor activity of the insulin-like growth factor-I receptor kinase inhibitor NVP-AEW541 in musculoskeletal tumors. Cancer Res. 2005, 65, 3868–3876. [Google Scholar] [CrossRef] [Green Version]
- Bitelman, C.; Sarfstein, R.; Sarig, M.; Attias-Geva, Z.; Fishman, A.; Werner, H.; Bruchim, I. IGF1R-directed targeted therapy enhances the cytotoxic effect of chemotherapy in endometrial cancer. Cancer Lett. 2013, 335, 153–159. [Google Scholar] [CrossRef] [PubMed]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shibel, R.; Sarfstein, R.; Nagaraj, K.; Lapkina-Gendler, L.; Laron, Z.; Dixit, M.; Yakar, S.; Werner, H. The Olfactory Receptor Gene Product, OR5H2, Modulates Endometrial Cancer Cells Proliferation via Interaction with the IGF1 Signaling Pathway. Cells 2021, 10, 1483. https://doi.org/10.3390/cells10061483
Shibel R, Sarfstein R, Nagaraj K, Lapkina-Gendler L, Laron Z, Dixit M, Yakar S, Werner H. The Olfactory Receptor Gene Product, OR5H2, Modulates Endometrial Cancer Cells Proliferation via Interaction with the IGF1 Signaling Pathway. Cells. 2021; 10(6):1483. https://doi.org/10.3390/cells10061483
Chicago/Turabian StyleShibel, Rand, Rive Sarfstein, Karthik Nagaraj, Lena Lapkina-Gendler, Zvi Laron, Manisha Dixit, Shoshana Yakar, and Haim Werner. 2021. "The Olfactory Receptor Gene Product, OR5H2, Modulates Endometrial Cancer Cells Proliferation via Interaction with the IGF1 Signaling Pathway" Cells 10, no. 6: 1483. https://doi.org/10.3390/cells10061483