Characterization of Apis mellifera Gastrointestinal Microbiota and Lactic Acid Bacteria for Honeybee Protection—A Review
Abstract
:1. Introduction
2. Apis mellifera Characterization
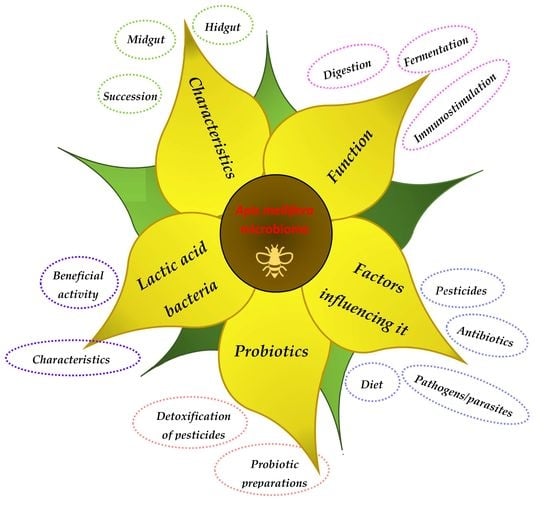
3. Honeybee Microbiota
3.1. Characteristics
3.2. Functions
3.3. Factors Affecting Honeybee Microbiota
4. LAB as a Significant Component of the Honeybee Microbiota and Their Beneficial Activities
5. Probiotics for Honeybees
| Strain | Pesticide/s | Effect | Reference |
|---|---|---|---|
| Human gut microbiota plus L. plantarum ATCC 11095 | Phoxim, chlorpyrifos, imidacloprid, thiamethoxam, emamectin benzoate, chlorpyrifos-d10, thiamethoxam-d4 | Metabolism of pesticides in the colon digests. The rate of the metabolism was significantly increased in the presence of L. plantarum. The strain reduced the relative amounts of six pesticides by 11.40–86.51%. | [204] |
| 282 LAB strains, L. plantarum RS60 and P. acidilactici D15 selected as the most efficient | Cypermethrin | 229 LAB strains removed the pesticide by at least 81% (binding), and 56% of cypermethrin was removed within 15 min by L. plantarum RS60 and P. acidilactici D15. No metabolites were detected. | [203] |
| L. plantarum LB-1 and LB-2 | Chlorpyrifos, deltamethrin | Degradation reached values of up to 96%. Metabolism of these insecticides was conducted by the esterase enzyme. Tested LAB used these compounds as carbon and energy sources. | [205] |
| P. acidilactici PA CNCM MA18/5 M | Thiamethoxam, boscalid | Tested pesticides deregulated genes involved in detoxification system (glutathione peroxidase-like 2, catalase) in honeybees. The strain abolished the harmful effects. | [193] |
| Ent. faecium E86, L. lactis subsp. lactis ATCC 11454; L. rhamnosus GG; Leuconostoc lactis ATCC 19256; L. mesenteroides subsp. mesenteroides ATCC 8293, P. pentosaceus ATCC 43200 | Chlorpyrifos | All LAB degraded chlorpyrifos by a minimum of 80.3%. In the case of P. pentosaceus, complete degradation was observed (below detection limit). | [206] |
| L. acidophilus, L. delbrueckii subsp. bulgaricus, L. plantarum, L. rhamnosus, L. casei, S. thermophilus, Bifidobacterium bifidum used as starter cultures | Organochlorine pesticide mixture (α-HCH, HCB, γ-HCH, g-chlordane, α-chlordane) | The starters contributed to a significant reduction in pesticide level during the production of yogurt and cheese. | [207] |
| 121 strains of L. plantarum, of which six with the highest activity were selected | Dimethoate, phorate, omethoate | All pesticides were degraded with different effectiveness depending on the strain—with omethoate, by up to 13%; phorate, by up to 36%; and dimethoate, by up to 27%. | [208] |
| L. plantarum ATCC 14917 | Imidacloprid | LAB reduced susceptibility to infection with honeybee pathogen S. marcescens Db11 in an insect model of D. melanogaster by immune-deficiency pathway. LAB did not bind or metabolize imidacloprid. | [113] |
| L. casei WYS3 | Chlorpyrifos | Viable pour culture bound 33.3–42% of exogenously added chlorpyrifos; acid-treated cells and heat-treated cells bound 32.0% and 77.2% chlorpyrifos, respectively. During rice straw silage fermentation, the reduction of chlorpyrifos was up to 72.0%. | [209] |
| L. rhamnosus GG (LGG), L. rhamnosus GR-1 (LGR-1) | Parathion, chlorpyrifos | Metabolism and passive binding of both pesticides by alive and heat-killed strains. Bacteria also reduced intestinal absorption of these compounds via Caco-2 Transwell model of the small intestine. | [210] |
| L. casei | Diazinon | Decrease of cytotoxicity of diazinon after treatment of HUVEC cells (human umbilical vein endothelial) with cell-free supernatant in a dose-dependent manner by nearly 51%. | [211] |
| L. plantarum BJ0021 | Endosulfan | Protective effect of LAB, which reduced toxicity of endosulfan in pregnant Wistar rats by amelioration of blood and urine biochemical values, and decrease in apoptosis of liver and kidney cells. | [212] |
| 10 LAB strains in skimmed milk (L. plantarum, L. helveticus, L. brevis, L. bulgaricus, L. lactis, Streptococcus thermophilus) | Chlorpyrifos, diazinon, fenitrothion, malathion, methyl parathion | Degradation of pesticides during fermentation of milk. The metabolism was conducted by LAB phosphatase enzymes. Different combinations of strains reduced the pesticide content to a greater extent than single strains. | [213] |
| L. plantarum DSMZ 20174 | Pirimiphos-methyl | Degradation of pesticide with 81% effectiveness during wheat fermentation without toxic effect on growth and activity of the strain. | [214] |
| L. fermentum MTCC 903, L. lactis MTCC 4185 | Chlorpyrifos | L. lactis and L. fermentum degraded chlorpyrifos to different metabolic end products—chlorpyrifos-oxon (in 61%) and 3,5,6-trichloro-2-pyridinol (in 70%), respectively. | [215] |
| L. brevis WCP902 | Chlorpyrifos | Complete degradation of the pesticide. Authors isolated a gene (opdB) encoding an organophosphorus hydrolase enzyme (OpdB) responsible for the degradation. | [216] |
| L. mesenteroides WCP907, L. brevis WCP902, L. plantarum WCP931, L. sakei WCP904 | Chlorpyrifos, coumaphos, diazinon, parathion, methylparathion | All compounds were utilized as the sole source of carbon and phosphorus during the fermentation of kimchi. Chlorpyrifos was degraded up to 100% within 9 days. Remaining pesticides were degraded by up to 82% within 12 days. | [217] |
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Villalba, A.; Maggi, M.; Ondarza, P.M.; Szawarski, N.; Miglioranza, K.S.B. Influence of land use on chlorpyrifos and persistent organic pollutant levels in honey bees. Bee bread and honey: Beehive exposure assessment. Sci. Total Environ. 2020, 713, 136554. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, P.; Moran, N.A. Functional and evolutionary insights into the simple yet specific gut microbiota of the honey bee from metagenomic analysis. Gut Microbes 2013, 4, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Ricigliano, V.A. Honey bee gut dysbiosis: A novel context of disease ecology. Curr. Opin. Insect Sci. 2017, 22, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Anjum, S.I.; Shah, A.H.; Aurongzeb, M.; Kori, J.; Azim, M.K.; Ansari, M.J.; Bin, L. Characterization of gut bacterial flora of Apis mellifera from north-west Pakistan. Saudi J. Biol. Sci. 2018, 25, 388–392. [Google Scholar] [CrossRef]
- Bonilla-Rosso, G.; Engel, P. Functional roles and metabolic niches in the honey bee gut microbiota. Curr. Opin. Microbiol. 2018, 43, 69–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bommuraj, V.; Chen, Y.; Birenboim, M.; Barel, S.; Shimshoni, J.A. Concentration- and time-dependent toxicity of commonly encountered pesticides and pesticide mixtures to honeybees (Apis mellifera L.). Chemosphere 2021, 266, 128974. [Google Scholar] [CrossRef] [PubMed]
- Hung, K.J.; Kingston, J.M.; Albrecht, M.; Holway, D.A.; Kohn, J.R. The worldwide importance of honey bees as pollinators in natural habitats. Proc. Bio. Sci. 2018, 285, 20172140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matilla, H.R.; Harris, J.L.; Otis, G.W. Timing of production of winter bees in honey bee (Apis mellifera) colonies. Insectes Sociaux 2001, 48, 88–93. [Google Scholar] [CrossRef]
- Amdam, G.V.; Omholt, S.W. The Regulatory Anatomy of Honeybee Lifespan. J. Theor. Biol. 2002, 216, 209–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardoso-Júnior, C.A.M.; Guidugli-Lazzarini, K.R.; Hartfelder, K. DNA methylation affects the lifespan of honey bee (Apis mellifera L.) workers – evidence for a regulatory module that involves vitellogenin expression but is independent of juvenile hormone function. Insect Biochem. Mol. Biol. 2018, 92, 21–29. [Google Scholar] [CrossRef]
- Han, F.; Wallberg, A.; Webster, M.T. From where did the Western honeybee (Apis mellifera) originate? Ecol. Evol. 2012, 2, 1949–1957. [Google Scholar] [CrossRef]
- Gupta, R.K. Chapter 2. Taxonomy and Distribution of Different Honeybee Species. In Beekeeping for Poverty Alleviation and Livelihood Security; Gupta, R.K., Reybroeck, W., van Veen, J.W., Gupta, A., Eds.; Springer Nature: Berlin, Germany, 2014; Volume 1. [Google Scholar]
- Glenny, W.; Cavigli, I.; Daughenbaugh, K.F.; Radford, R.; Kegley, S.E.; Flenniken, M.L. Honey bee (Apis mellifera) colony health and pathogen composition in migratory beekeeping operations involved in California almond pollination. PLoS ONE 2017, 12, e0182814. [Google Scholar] [CrossRef]
- Hewlett, S.E.; Wareham, D.M.; Barron, A.B. Honey bee (Apis mellifera) sociability and nestmate affiliation are dependent on the social environment experienced post-eclosion. J. Exp. Biol. 2017, 221, eb173054. [Google Scholar] [CrossRef] [Green Version]
- Amiri, E.; Strand, M.K.; Rueppell, O.; Tarpy, D.R. Queen quality and the impact of honey bee diseases on queen health: Potential for interactions between two major threats to colony health. Insects 2017, 8, 48. [Google Scholar] [CrossRef]
- Lee, K.V.; Goblirsch, M.; McDermott, E.; Tarpy, D.R.; Spivak, M. Is the brood pattern within a honey bee colony a reliable indicator of queen quality? Insects 2019, 10, 12. [Google Scholar] [CrossRef] [Green Version]
- Negri, P.; Villalobos, E.; Szawarski, N.; Damiani, N.; Gende, L.; Garrido, M.; Maggi, M.; Quintana, S.; Lamattina, L.; Eguaras, M. Towards Precision Nutrition: A Novel Concept Linking Phytochemicals, Immune Response and Honey Bee Health. Insects 2019, 10, 401. [Google Scholar] [CrossRef] [Green Version]
- Iorizzo, M.; Pannella, G.; Lombardi, S.J.; Ganassi, S.; Testa, B.; Succi, M.; Sorrentino, E.; Petrarca, S.; De Cristofaro, A.; Coppola, R.; et al. Inter- and intra-species diversity of lactic acid bacteria in Apis mellifera ligustica colonies. Microorganisms 2020, 8, 1578. [Google Scholar] [CrossRef]
- Brodschneider, R.; Crailsheim, K. Nutrition and health in honey bees. Apidologie 2010, 41, 278–294. [Google Scholar] [CrossRef]
- Danner, N.; Keller, A.; Härtel, S.; Steffan-Dewenter, I. Honey bee foraging ecology: Season but not landscape diversity shapes the amount and diversity of collected pollen. PLoS ONE 2017, 12, e0183716. [Google Scholar] [CrossRef] [Green Version]
- Hoover, S.E.; Ovinge, L.P. Pollen Collection, Honey Production, and Pollination Services: Managing Honey Bees in an Agricultural Setting. J. Econ. Entomol. 2018, 111, 1509–1516. [Google Scholar] [CrossRef] [PubMed]
- Vieira, K.I.C.; Azevedo Werneck, H.; Santos Júnior, J.E.; Silva Flores, D.S.; Serrão, J.E.; Campos, L.A.D.O.; Resende, H.C. Bees and the environmental impact of the rupture of the fundão dam. Integr. Environ. Assess. Manag. 2020, 16, 631–635. [Google Scholar] [CrossRef] [PubMed]
- Carroll, M.J.; Brown, N.; Goodall, C.; Downs, A.M.; Sheenan, T.H.; Anderson, K.E. Honey bees preferentially consume freshly stored pollen. PLoS ONE 2017, 12, e0175933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, G.A.; Nicolson, S.W.; Shafir, S. Nutritional physiology and ecology of honey bees. Annu. Rev. Entomol. 2018, 63, 327–344. [Google Scholar] [CrossRef]
- Nuru, A.; Hepburn, H.R. Pollen grains of some bee plants of Ethiopia. In Proceedings of the 37th International Apicultural Congress, Durban, South Africa, 28 October–1 November 2001. [Google Scholar]
- Bertazzini, M.; Medrzycki, P.; Bortolotti, L.; Maistrello, L.; Forlani, G. Amino acid content and nectar choice by forager honeybees (Apis mellifera L.). Amino Acids 2010, 39, 315–318. [Google Scholar] [CrossRef] [Green Version]
- Nicolson, S.W. Bee food: The chemistry and nutritional value of nectar, pollen and mixtures of the two. Afr. Zool. 2011, 46, 197–204. [Google Scholar] [CrossRef]
- Cornara, L.; Biagi, M.; Xiao, J.; Burlando, B. Therapeutic properties of bioactive compounds from different honeybee products. Front. Pharmacol. 2017, 8. [Google Scholar] [CrossRef]
- Kurek-Górecka, A.; Górecki, M.; Rzepecka-Stojko, A.; Balwierz, R.; Stojko, J. Bee products in dermatology and skin care. Molecules 2020, 25, 556. [Google Scholar] [CrossRef] [Green Version]
- Ball, D.W. The chemical composition of honey. J. Chem. Edu. 2007, 84, 1643. [Google Scholar] [CrossRef]
- Svečnjak, L.; Prđun, S.; Rogina, J.; Bubalo, D.; Jerković, I. Characterization of Satsuma mandarin (Citrus unshiu Marc.) nectar-to-honey transformation pathway using FTIR-ATR spectroscopy. Food Chem. 2017, 232, 286–294. [Google Scholar] [CrossRef]
- Vezeteu, T.V.; Bobiş, O.; Moritz, R.F.A.; Buttstedt, A. Food to some. poison to others—honeybee royal jelly and its growth inhibiting effect on European Foulbrood bacteria. MicrobiologyOpen 2016, 6, e00397. [Google Scholar] [CrossRef] [Green Version]
- Melliou, E.; Chinou, I. Chapter 8—Chemistry and bioactivities of royal jelly. In Studies in Natural Products Chemistry; Elsevier: Amsterdam, The Netherlands, 2014; Volume 43, pp. 261–290. [Google Scholar] [CrossRef]
- Mannoor, M.; Shimabukuro, I.; Tsukamotoa, M.; Watanabe, H.; Yamaguchi, K.; Sato, Y. Honeybee royal jelly inhibits autoimmunity in SLE-prone NZB × NZW F1 mice. Lupus 2009, 18, 44–52. [Google Scholar] [CrossRef]
- Anderson, K.E.; Ricigliano, V.A.; Mott, B.M.; Copeland, D.C.; Floyd, A.S.; Maes, P. The queen’s gut refines with age: Longevity phenotypes in a social insect model. Microbiome 2018, 6, 108. [Google Scholar] [CrossRef] [Green Version]
- Salazar-Olivo, L.A.; Paz-González, V. Screening of biological activities present in honeybee (Apis mellifera) royal jelly. Toxicol. In Vitro 2005, 19, 645–651. [Google Scholar] [CrossRef]
- Pascale, A.; Marchesi, N.; Marelli, C.; Coppola, A.; Luzi, L.; Govoni, S.; Gazzaruso, C. Microbiota and metabolic diseases. Endocrine 2018, 61, 357–371. [Google Scholar] [CrossRef]
- Greiner, T.; Bäckhed, F. Effects of the gut microbiota on obesity and glucose homeostasis. Trends Endocrinol. Metab. 2011, 22, 117–123. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef]
- Gagliardi, A.; Totino, V.; Cacciotti, F.; Iebba, V.; Neroni, B.; Bonfiglio, G.; Trancassini, M.; Passariello, C.; Pantanella, F.; Schippa, S. Rebuilding the gut microbiota ecosystem. Int. J. Environ. Res. Public Health 2018, 15, 1679. [Google Scholar] [CrossRef] [Green Version]
- Agus, A.; Planchais, J.; Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [Green Version]
- Molloy, M.J.; Bouladoux, N.; Belkaid, Y. Intestinal microbiota: Shaping local and systemic immune responses. Semin. Immunol. 2012, 24, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Belkaid, Y.; Harrison, O.J. Homeostatic Immunity and the Microbiota. Immunity 2017, 46, 562–576. [Google Scholar] [CrossRef] [Green Version]
- Biedermann, L.; Rogler, G. The intestinal microbiota: Its role in health and disease. Eur. J. Pediatr. 2015, 174, 151–167. [Google Scholar] [CrossRef]
- El Aidy, S.; Dinan, T.G.; Cryan, J.F. Gut microbiota: The conductor in the orchestra of immune–neuroendocrine communication. Clin. Ther. 2015, 37, 954–967. [Google Scholar] [CrossRef]
- Mu, C.; Yang, Y.; Zhu, W. Gut Microbiota: The Brain Peacekeeper. Front. Microbiol. 2016, 7, 345. [Google Scholar] [CrossRef] [Green Version]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Steele, M.I.; Leonard, S.P.; Motta, E.V.S.; Moran, N.A. Honey bees as models for gut microbiota research. Lab. Animal. 2018, 47, 317–325. [Google Scholar] [CrossRef]
- Dong, Z.X.; Li, H.Y.; Chen, Y.F.; Wang, F.; Deng, X.Y.; Lin, L.B.; Zhang, Q.L.; Li, J.L.; Guo, J. Colonization of the gut microbiota of honey bee (Apis mellifera) workers at different developmental stages. Microbiol. Res. 2020, 231, 126370. [Google Scholar] [CrossRef]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef] [Green Version]
- Motta, E.V.S.; Raymann, K.; Moran, N.A. Glyphosate perturbs the gut microbiota of honey bees. Proc. Natl. Acad. Sci. 2018, 115, 10305–10310. [Google Scholar] [CrossRef] [Green Version]
- Bleau, N.; Bouslama, S.; Giovenazzo, P.; Derome, N. Dynamics of the honeybee (Apis mellifera) gut microbiota throughout the overwintering period in Canada. Microorganisms 2020, 29, 1146. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Evans, J.D.; Li, W.F.; Zhao, Y.Z.; DeGrandi-Hoffman, G.; Huang, S.K.; Li, Z.G.; Hamilton, M.; Chen, Y.P. New evidence showing that the destruction of gut bacteria by antibiotic treatment could increase the honey bee’s vulnerability to Nosema infection. PLoS ONE 2017, 12, e0187505. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of Acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. 2012, 109, 11002–11007. [Google Scholar] [CrossRef] [Green Version]
- Vásquez, A.; Forsgren, E.; Fries, I.; Paxton, R.J.; Flaberg, E.; Szekely, L.; Olofsson, T.C. Symbionts as major modulators of insect health: Lactic acid bacteria and honeybees. PLoS ONE 2012, 7, e33188. [Google Scholar] [CrossRef]
- Corby-Harris, V.; Snyder, L.A.; Schwan, M.R.; Maes, P.; McFrederick, Q.S.; Anderson, K.E. Origin and effect of Alpha 2.2 Acetobacteraceae in honey bee larvae and description of Parasaccharibacter apium gen. nov., sp. nov. Appl. Environ. Microbiol. 2014, 80, 7460–7472. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.C.; Fruciano, C.; Hildebrand, F.; Al Toufalilia, H.; Balfour, N.J.; Bork, P.; Engel, P.; Ratnieks, F.L.W.; Hughes, W.O. Gut microbiota composition is associated with environmental landscape in honey bees. Ecol. Evol. 2017, 8, 441–451. [Google Scholar] [CrossRef] [Green Version]
- Alberoni, D.; Gaggìa, F.; Baffoni, L.; Di Gioia, D. Beneficial microorganisms for honey bees: Problems and progresses. App. Microbiol. Biotechnol. 2016, 100, 9469–9482. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Moran, N.A.; Hansen, A.K.; Powell, J.E.; Sabree, Z.L. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE 2020, 7, e36393. [Google Scholar] [CrossRef] [Green Version]
- Tola, Y.H.; Waweru, J.W.; Hurst, G.D.D.; Slippers, B.; Paredes, J.C. Characterization of the Kenyan honey bee (Apis mellifera) gut microbiota: A first look at tropical and Sub-Saharan African bee associated microbiomes. Microorganisms 2020, 8, 1721. [Google Scholar] [CrossRef]
- Kešnerová, L.; Emery, O.; Troilo, M.; Liberti, J.; Erkosar, B.; Engel, P. Gut microbiota structure differs between honeybees in winter and summer. ISME J. 2020, 14, 801–814. [Google Scholar] [CrossRef] [Green Version]
- Kešnerová, L.; Moritz, R.; Engel, P. Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 2016, 66, 414–421. [Google Scholar] [CrossRef]
- Hilgarth, M.; Redwitz, J.; Ehrmann, M.A.; Vogel, R.F.; Jakob, F. Bombella favorum sp. nov. and Bombella mellum sp. nov., two novel species isolated from the honeycombs of Apis mellifera. Int. J. Syst. Evol. Microbiol. 2021. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Apibacter adventoris gen. nov., sp. nov., a member of the phylum Bacteroidetes isolated from honey bees. Int. J. Syst. Evol. Microbiol. 2016, 66, 1323–1329. [Google Scholar] [CrossRef]
- Kwong, W.K.; Steele, M.I.; Moran, N.A. Genome sequences of Apibacter spp., gut symbionts of Asian honey bees. Genome Biol. Evol. 2018, 10, 1174–1179. [Google Scholar] [CrossRef]
- Subotic, S.; Boddicker, A.M.; Nguyen, V.M.; Rivers, J.; Briles, C.E.; Mosier, A.C. Honey bee microbiome associated with different hive and sample types over a honey production season. PLoS ONE 2019, 14, e0223834. [Google Scholar] [CrossRef]
- Wang, S.; Wang, L.; Fan, X.; Yu, C.; Feng, L.; Yi, L. An insight into diversity and functionalities of gut microbiota in insects. Curr. Microbiol. 2020, 77, 1976–1986. [Google Scholar] [CrossRef]
- Wang, H.; Liu, C.; Liu, Z.; Wan, Y.; Ma, L.; Xu, B. The different dietary sugars modulate the composition of the gut microbiota in honeybee during overwintering. BMC Microbiol. 2020, 20, 61. [Google Scholar] [CrossRef]
- Pernice, M.; Simpson, S.J.; Ponton, F. Towards an integrated understanding of gut microbiota using insects as model systems. J. Insect Physiol. 2014, 69, 12–18. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects – diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Cianci, R.; Pagliari, D.; Piccirillo, C.A.; Fritz, J.H.; Gambassi, G. The microbiota and immune system crosstalk in health and disease. MediatorsInflamm 2018, 2018, 2912539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, W.; Syrenne, R.; Sun, J.-Z.; Yuan, J.S. Molecular approaches to study the insect gut symbiotic microbiota at the “omics” age. Insect Sci. 2010, 17, 199–219. [Google Scholar] [CrossRef]
- Jing, T.-Z.; Qi, F.-H.; Wang, Z.-Y. Most dominant roles of insect gut bacteria: Digestion. detoxification or essential nutrient provision? Microbiome 2020, 8. [Google Scholar] [CrossRef] [Green Version]
- Azambuja, P.; Garcia, E.S.; Ratcliffe, N.A. Gut microbiota and parasite transmission by insect vectors. Trends in Parasitology 2005, 21, 568–572. [Google Scholar] [CrossRef]
- Cirimotich, C.M.; Ramirez, J.L.; Dimopoulos, G. Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe 2011, 10, 307–310. [Google Scholar] [CrossRef] [Green Version]
- Silva, M.S.; Rabadzhiev, Y.; Renon Eller, M.; Iliev, I.; Ivanova, I.; Santana, W.C. Microorganisms in honey. Honey analysis. IntechOpen 2017, S233–S258. [Google Scholar] [CrossRef] [Green Version]
- Pachila, A.; Ptaszyńska, A.A.; Wicha, M.; Oleńska, E.; Małek, W. Fascinating fructophilic lactic acid bacteria associated with various fructose-rich niches. Ann. Univ. Mariae Curie Sklodowska Med. 2017, 72, S41–S50. [Google Scholar] [CrossRef]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef] [Green Version]
- Egert, M.; Simmering, R. The microbiota of the human skin. advances in experimental medicine and biology. Adv. Exp. Med. Biol. 2016, 90, 61–81. [Google Scholar] [CrossRef]
- Schroeder, B.O.; Bäckhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nature Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.C.; Kim, S.H.; You, H.; Kim, B.; Kim, A.C.; Lee, K.A.; Yoon, J.H.; Ryu, J.H.; Lee, W.J. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 2011, 4, 334–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purchiaroni, F.; Tortora, A.; Gabrielli, M.; Bertucci, F.; Gigange, G.; Laniro, G.; Ojetti, V.; Scarpellini, E.; Gasbarrini, A. The role of intestinal microbiota and the immune system. Eur. Rev. Med. Pharmacol. Sci. 2013, 17, 323–333. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [Green Version]
- Valentini, M.; Piermattei, A.; Di Sante, G.; Migliara, G.; Delogu, G.; Ria, F. Immunomodulation by gut microbiota: Role of toll-like receptor expressed by T cells. J. Immunol. Res. 2014, 586939, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Yiu, J.H.C.; Dorweiler, B.; Woo, C.W. Interaction between gut microbiota and toll-like receptor: From immunity to metabolism. J. Mol. Med. 2016, 95, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Cederlund, A.; Gudmundsson, G.H.; Agerberth, B. Antimicrobial peptides important in innate immunity. FEBS J. 2011, 78, 3942–3951. [Google Scholar] [CrossRef]
- Westfall, S.; Lomis, N.; Prakash, S. Longevity extension in Drosophila through gut-brain communication. Sci. Rep. 2018, 8, 8362. [Google Scholar] [CrossRef]
- Leger, L.; McFrederick, Q.S. The gut–brain–microbiome axis in bumble bees. Insects 2020, 11, 517. [Google Scholar] [CrossRef]
- Liberti, J.; Engel, P. The gut microbiota—brain axis of insects. Curr. Opin. Insect Sci. 2020, 39, 6–13. [Google Scholar] [CrossRef]
- Harris, J.W.; Woodring, J. Effects of stress, age, season, and source colony on levels of octopamine, dopamine and serotonin in the honey bee (Apis mellifera L.) brain. J. Ins. Physiol. 1992, 38, 29–35. [Google Scholar] [CrossRef]
- Vernier, C.L.; Chin, I.M.; Adu-Oppong, B.; Krupp, J.J.; Levine, J.; Dantas, G.; Ben-Shahar, Y. The gut microbiome defines social group membership in honey bee colonies. Sci. Adv. 2020, 6, eabd3431. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vergnolle, N. Protease inhibition as new therapeutic strategy for GI diseases. Gut 2016, 65, 1215–1224. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Zheng, Y.; Chen, Y.; Wang, S.; Chen, Y.; Hu, F.; Zheng, H. Honey bee (Apis mellifera) gut microbiota promotes host endogenous detoxification capability via regulation of P450 gene expression in the digestive tract. Microb. Biotechnol. 2020, 13, 1201–1212. [Google Scholar] [CrossRef]
- Van Engelsdorp, D.; Traynor, K.S.; Andree, M.; Lichtenberg, E.M.; Chen, Y.; Saegerman, C.; Cox-Foster, D.L. Colony Collapse Disorder (CCD) and bee age impact honey bee pathophysiology. PLoS ONE 2017, 12, e0179535. [Google Scholar] [CrossRef] [Green Version]
- Sommer, F.; Bäckhed, F. The gut microbiota—Masters of host development and physiology. Nature Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Muñoz-Colmenero, M.; Baroja-Careaga, I.; Kovačić, M.; Filipi, J.; Puškadija, Z.; Kezić, N.; Estonba, A.; Büchler, R.; Zarraonaindia, I. Differences in honey bee bacterial diversity and composition in agricultural and pristine environments—A field study. Apidologie 2020, 51, 1018–1037. [Google Scholar] [CrossRef]
- Fisher, A.; DeGrandi-Hoffman, G.; Smith, B.H.; Johnson, M.; Kaftanoglu, O.; Cogley, T.; Fewell, J.H.; Harrison, J.F. Colony field test reveals dramatically higher toxicity of a widely used mito-toxic fungicide on honey bees (Apis mellifera). Environ. Pollut. 2021, 269, 115964. [Google Scholar] [CrossRef]
- Flores, J.M.; Gámiz, V.; Gil-Lebrero, S.; Rodríguez, I.; Navas, F.J.; García-Valcárcel, A.I.; Cutillas, V.; Fernández-Alba, A.R.; Hernando, M.D. A three-year large scale study on the risk of honey bee colony exposure to blooming sunflowers grown from seeds treated with thiamethoxam and clothianidin neonicotinoids. Chemosphere 2021, 262, 127735. [Google Scholar] [CrossRef]
- Milone, J.P.; Tarpy, D.R. Effects of developmental exposure to pesticides in wax and pollen on honey bee (Apis mellifera) queen reproductive phenotypes. Sci. Rep. 2021, 11, 1020. [Google Scholar] [CrossRef]
- Miotelo, L.; Mendes Dos Reis, A.L.; Malaquias, J.B.; Malaspina, O.; Roat, T.C. Apis mellifera and Melipona scutellaris exhibit differential sensitivity to thiamethoxam. Environ. Pollut. 2021, 268, 115770. [Google Scholar] [CrossRef]
- Kakumanu, M.L.; Reeves, A.M.; Anderson, T.D.; Rodrigues, R.R.; Williams, M.A. Honey bee gut microbiome is altered by in-hive pesticide exposures. Front. Microbiol. 2016, 7, 1255. [Google Scholar] [CrossRef] [Green Version]
- Rouzé, R.; Moné, A.; Delbac, F.; Belzunces, L.; Blot, N. The honeybee gut microbiota is altered after chronic exposure to different families of insecticides and infection by Nosema ceranae. Microbes Environ. 2019, 34, 226–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motta, E.; Mak, M.; De Jong, T.K.; Powell, J.E.; O’Donnell, A.; Suhr, K.J.; Riddington, I.M.; Moran, N.A. Oral or Topical Exposure to Glyphosate in Herbicide Formulation Impacts the Gut Microbiota and Survival Rates of Honey Bees. Appl. Environ. Microbiol. 2020, 86, e01150-20. [Google Scholar] [CrossRef]
- Liu, Y.J.; Qiao, N.H.; Diao, Q.Y.; Jing, Z.; Vukanti, R.; Dai, P.L.; Ge, Y. Thiacloprid exposure perturbs the gut microbiota and reduces the survival status in honeybees. J. Hazard. Mater. 2020, 5, 389. [Google Scholar] [CrossRef]
- Zhu, L.; Qi, S.; Xue, X.; Niu, X.; Wu, L. Nitenpyram disturbs gut microbiota and influences metabolic homeostasis and immunity in honey bee (Apis mellifera L.). Environ Pollut. 2020, 258, 113671. [Google Scholar] [CrossRef]
- Alberoni, D.; Favaro, R.; Baffoni, L.; Angeli, S.; Di Gioia, D. Neonicotinoids in the agroecosystem: In-field long-term assessment on honeybee colony strength and microbiome. Sci. Total. Environ. 2021, 15, 762. [Google Scholar] [CrossRef]
- Daisley, B.A.; Trinder, M.; McDowell, T.W.; Welle, H.; Dube, J.S.; Ali, S.N.; Leong, H.S.; Sumarah, M.W.; Reid, G. Neonicotinoid-induced pathogen susceptibility is mitigated by Lactobacillus plantarum immune stimulation in a Drosophila melanogaster model. Sci. Rep. 2017, 7, 2703. [Google Scholar] [CrossRef]
- Diaz, T.; Del-Val, E.; Ayala, R.; Larsen, J. Alterations in honey bee gut microorganisms caused by Nosema spp. and pest control methods. Pest. Manag. Sci. 2019, 75, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.A.; Ansari, M.J.; Al-Ghamdi, A.; Nuru, A.; Harakeh, S.; Iqbal, J. Investigation of gut microbial communities associated with indigenous honey bee (Apis mellifera jemenitica) from two different eco-regions of Saudi Arabia. Saudi J. Biol. Sci. 2017, 24, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Castelli, L.; Branchiccela, B.; Garrido, M.; Invernizzi, C.; Porrini, M.; Romero, H.; Santos, E.; Zunino, P.; Antúnez, K. Impact of nutritional stress on honeybee gut microbiota, immunity, and Nosema ceranae infection. Microb. Ecol. 2020, 80, 908–919. [Google Scholar] [CrossRef]
- Saelao, P.; Borba, R.S.; Ricigliano, V.; Spivak, M.; Simone-Finstrom, M. Honeybee microbiome is stabilized in the presence of propolis. Biol. Lett. 2020, 16, 20200003. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, Z.; Chen, X.; Hong, Z.; Lin, W.; Mu, X.; Hu, X.; Zheng, H. High-Fat Diets with Differential Fatty Acids Induce Obesity and Perturb Gut Microbiota in Honey Bee. Int. J. Mol. Sci. 2021, 22, 834. [Google Scholar] [CrossRef]
- Daisley, B.A.; Pitek, A.P.; Chmiel, J.A.; Gibbons, S.; Chernyshova, A.M.; Al, K.F.; Faragalla, K.M.; Burton, J.P.; Thompson, G.J.; Reid, G. Lactobacillus spp. attenuate antibiotic-induced immune and microbiota dysregulation in honey bees. Commun. Biol. 2020, 3, 534. [Google Scholar] [CrossRef]
- Ludvigsen, J.; Porcellato, D.; L’Abée-Lund, T.M.; Amdam, G.V.; Rudi, K. Geographically widespread honeybee-gut symbiont subgroups show locally distinct antibiotic-resistant patterns. Mol. Ecol. 2017, 26, 6590–6607. [Google Scholar] [CrossRef]
- Reybroeck, W. Residues of antibiotics and chemotherapeutics in honey. J. Api. Res. 2017, 57, 97–112. [Google Scholar] [CrossRef]
- Raymann, K.; Bobay, L.M.; Moran, N.A. Antibiotics reduce genetic diversity of core species in the honeybee gut microbiome. Mol. Ecol. 2018, 27, 2057–2066. [Google Scholar] [CrossRef]
- Ortiz-Alvarado, Y.; Clark, D.R.; Vega-Melendez, C.J.; Flores-Cruz, Z.; Domingez-Bello, M.G.; Giray, T. Antibiotics in hives an their effect on honey bee physiology and behavioral development. Biol. Open 2020, 9, bio053884. [Google Scholar] [CrossRef]
- Blaser, M.J. Antibiotic use and its consequences for the normal microbiome. Science 2016, 352, 544–545. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.; Armstrong, T.N. Inhibition of the American foulbrood bacterium, Paenibacillus larvae, by bacteria isolated from honey bees. J. Apic. Res. 2005, 44, 168–171. [Google Scholar] [CrossRef]
- Evans, J.D.; Spivak, M. Socialized medicine: Individual and communal disease barriers in honey bees. J. Invertebr. Pathol. 2010, 103, S62–S72. [Google Scholar] [CrossRef]
- Reybroeck, W.; Daeseleire, E.; De Brabander, H.F.; Herman, L. Antimicrobials in beekeeping. Vet. Microbiol. 2012, 158, 1–11. [Google Scholar] [CrossRef]
- Tian, B.; Fadhil, N.H.; Powell, J.E.; Kwong, W.K.; Moran, N.A. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 2012, 3, e00377-12. [Google Scholar] [CrossRef] [Green Version]
- Ludvigsen, J.; Amdam, G.V.; Rudi, K.; L’Abée-Lund, T.M. Detection and characterization of streptomycin resistance (strA-strB) in a honeybee gut symbiont (Snodgrassella alvi) and the associated risk of antibiotic resistance transfer. Microb. Ecol. 2018, 76, 588–591. [Google Scholar] [CrossRef]
- The European Green Deal. Communication from the Commission to the European Parliament, the European Council. In Proceedings of the European Economic and Social Committee and the Committee of the Regions, Brussels, Belgium, 11 December 2019.
- Regulation (EC) No 178/2002 of the European Parliament and of the Council of 28 January 2002 Laying down the General Principles and Requirements of Food Law, Establishing the European Food Safety Authority and Laying down Procedures in Matters of Food Safety, O.J.L. 31. 1 February 2002; 1–24.
- Commission Regulation (EU) No 415/2013 of 6 May 2013 Laying down Additional Responsibilities and Tasks for the EU Reference Laboratories for Rabies, Bovine Tuberculosis and Bee Health, Amending Regulation (EC) No 737/2008 and Repealing Regulation (EU) No 87/2011, O.J.L. 125. 7 May 2013; 7–12.
- Council Directive 2001/110/EC of 20 December 2001 Relating to Honey, O.J.L. 10. 12 January 2002; 47–52.
- Regulation (EC) No 470/2009 of the European Parliament and of the Council of 6 May 2009 Laying down Community Procedures for the Establishment of Residue Limits of Pharmacologically Active Substances in Foodstuffs of Animal Origin, Repealing Council Regulation (EEC) No 2377/90 and Amending Directive 2001/82/EC of the European Parliament and of the Council and Regulation (EC) No 726/2004 of the European Parliament and of the Council, O.J.L. 152. 16 June 2009; 11–22.
- Commission Regulation (EU) No 37/2010 of 22 December 2009 on Pharmacologically Active Substances and Their Classification Regarding Maximum Residue Limits in Foodstuffs of Animal Origin, O.J.L. 15. 20 January 2010; 1–72.
- Regulation (EU) 2019/6 of the European Parliament and of the Council of 11 December 2018 on Veterinary Medicinal Products and Repealing Directive 2001/82/EC, O.J.L. 4. 7 January 2019; 43–167.
- Mutlu, E.A.; Comba, I.Y.; Cho, T.; Engen, P.A.; Yazıcı, C.; Soberanes, S.; Hamanaka, R.B.; Niğdelioğlu, R.; Meliton, A.Y.; Ghio, A.J.; et al. Inhalational exposure to particulate matter air pollution alters the composition of the gut microbiome. Environ. Pollut. 2018, 240, 817–830. [Google Scholar] [CrossRef]
- Sampson, H.; Ketley, J.; Mallon, E.; Morrissey, J. Impact of air pollution on buff-tailed bumblebees (Bombus terrestris) and their gut microbiome. Access Microbiol. 2020, 2, 7A. [Google Scholar] [CrossRef]
- Costa, A.; Veca, M.; Barberis, M.; Tosti, A.; Notaro, G.; Nava, S.; Lazzari, M.; Agazzi, M.; Tangorra, F.M. Heavy metals on honeybees indicate their concentration in the atmosphere. a proof of concept. Ital. J. Anim. Sci. 2019, 18, 309–315. [Google Scholar] [CrossRef] [Green Version]
- Rothman, J.A.; Leger, L.; Kirkwood, J.S.; McFrederick, Q.S. Cadmium and Selenate Exposure Affects the Honey Bee Microbiome and Metabolome, and Bee-Associated Bacteria Show Potential for Bioaccumulation. Appl. Environ. Microbiol. 2019, 85, e01411-19. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, J.; Zhao, L.; Mu, X.; Wang, C.; Wang, M.; Xue, X.; Qi, S.; Wu, L. Gut microbiota protects honey bees (Apis mellifera L.) against polystyrene microplastics exposure risks. J. Hazard. Mater. 2021, 402, 123828. [Google Scholar] [CrossRef]
- Piccart, K.; Vásquez, A.; Piepers, S.; De Vliegher, S.; Olofsson, T.C. Short communication: Lactic acid bacteria from the honeybee inhibit the in vitro growth of mastitis pathogens. J. Dairy Sci. 2016, 99, 2940–2944. [Google Scholar] [CrossRef] [PubMed]
- Batista, V.L.; da Silva, T.F.; de Jesus, L.C.L.; Tapia-Costa, A.P.; Drumond, M.M.; Azevedo, V.; Mancha-Agresti, P. Lactic Acid Bacteria as Delivery Vehicle for Therapeutics Applications. Methods Mol Biol. 2021, 2183, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Perczak, A.; Goliński, P.; Bryła, M.; Waśkiewicz, A. The efficiency of lactic acid bacteria against pathogenic fungi and mycotoxins. Arh. Hig. Rada. Toksikol. 2018, 69, 32–45. [Google Scholar] [CrossRef] [Green Version]
- Mokoena, M.P. Lactic acid bacteria and their bacteriocins: Classification, biosynthesis and applications against uropathogens: A Mini-Review. Molecules 2017, 22, 1255. [Google Scholar] [CrossRef]
- Hatti-Kaul, R.; Chen, L.; Dishisha, T.; Enshasy, H.E. Lactic acid bacteria: From starter cultures to producers of chemicals. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [Green Version]
- Ouattara, D.H.; Ouattara, H.G.; Goualie, B.G.; Kouame, L.M.; Niamke, S.L. Biochemical and functional properties of lactic acid bacteria isolated from Ivorian cocoa fermenting beans. J. Appl. Biosci. 2014, 77, 6489–6499. [Google Scholar] [CrossRef]
- Blajman, J.E.; Páez, R.B.; Vinderola, C.G.; Lingua, M.S.; Signorini, M.L. A meta-analysis on the effectiveness of homofermentative and heterofermentative lactic acid bacteria for corn silage. J. Appl. Microbiol. 2018. [Google Scholar] [CrossRef]
- Mora-Villalobos, J.A.; Montero-Zamora, J.; Barboza, N.; Rojas-Garbanzo, C.; Usaga, J.; Redondo-Solano, M.; Schroedter, L.; Olszewska-Widdrat, A.; López-Gómez, J.P. Multi-Product Lactic Acid Bacteria Fermentations: A Review. Fermentation 2020, 6, 23. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Pang, H.; Zhang, H.; Cai, Y. Biodiversity of lactic acid bacteria. Lactic Acid Bacteria 2014, 103–203. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Giraffa, G. Selection and design of lactic acid bacteria probiotic cultures. Engineering in Life Sciences 2012, 12, 391–398. [Google Scholar] [CrossRef]
- Tanasupawat, S.; Okada, S.; Komagata, K. Lactic acid bacteria found in fermented fish in Thailand. J. Gen. Appl. Microbiol. 1998, 44, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Simova, E.; Beshkova, D.; Angelov, A.; Hristozova, T.; Frengova, G.; Spasov, Z. Lactic acid bacteria and yeasts in kefir grains and kefir made from them. J. Ind. Microbiol. Biotechnol. 2002, 28, 1–6. [Google Scholar] [CrossRef]
- Reina, L.D.; Breidt, F.; Fleming, H.P.; Kathariou, S. Isolation and selection of lactic acid bacteria as biocontrol agents for nonacidified, refrigerated pickles. J. Food Sci. 2005, 70, M7–M11. [Google Scholar] [CrossRef]
- Smit, G.; Smit, B.A.; Engels, W.J.M. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 2005, 29, 591–610. [Google Scholar] [CrossRef]
- Plengvidhya, V.; Breidt, F.; Lu, Z.; Fleming, H.P. DNA fingerprinting of lactic acid bacteria in sauerkraut fermentations. Appl. Environ. Microbiol. 2007, 73, 7697–7702. [Google Scholar] [CrossRef] [Green Version]
- Hurtado, A.; Reguant, C.; Bordons, A.; Rozès, N. Lactic acid bacteria from fermented table olives. Food Microbiol. 2012, 31, 1–8. [Google Scholar] [CrossRef]
- Bettache, G.; Fatma, A.; Miloud, H.M.; Mebrouk, K.M. Isolation and identification of lactic acid bacteria from Dhan, a traditional butter and their major technological traits guessas bettache. World J. Dairy Food Sci. 2013, 7, 101–108. [Google Scholar] [CrossRef]
- George, F.; Daniel, C.; Thomas, M.; Singer, E.; Guilbaud, A.; Tessier, F.J.; Revol-Junelles, A.M.; Borges, F.; Foligné, B. Occurrence and dynamism of lactic acid bacteria in distinct ecological niches: A multifaceted functional health perspective. Front. Microbiol. 2018, 9, 2899. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, D.S.; Moller, P.L.; Rosenfeldt, V.; Paerregaard, A.; Michaelsen, K.F.; Jakobsen, M. Case study of the distribution of mucosa-associated Bifidobacterium species, Lactobacillus species, and other lactic acid bacteria in the human colon. Appl. Environ. Microbiol. 2003, 69, 7545–7548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, L.; Tao, L.; Pavlova, S.I.; So, J.-S.; Kiwanuka, N.; Namukwaya, Z.; Saberbein, B.A.; Wawer, M. Species diversity and relative abundance of vaginal lactic acid bacteria from women in Uganda and Korea. J. Appl. Microbiol. 2006, 102, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Lombardi, S.J.; Ganassi, S.; Testa, B.; Ianiro, M.; Letizia, F.; Succi, M.; Tremonte, P.; Vergalito, F.; Cozzolino, A.; et al. Antagonistic activity against Ascosphaera apis and functional properties of Lactobacillus kunkeei strains. Antibiotics 2020, 9, 262. [Google Scholar] [CrossRef] [PubMed]
- Pessione, E. Lactic acid bacteria contribution to gut microbiota complexity: Lights and shadows. Front Cell Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef] [Green Version]
- Serna-Cock, L.; Rojas-Dorado, M.; Ordoñez-Artunduaga, D.; García-Salazar, A.; García-González, E.; Aguilar, C.N. Crude extracts of metabolites from co-cultures of lactic acid bacteria are highly antagonists of Listeria monocytogenes. Heliyon 2019, 5, e02448. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of lactic acid bacteria: Extending the family. Applied Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef] [Green Version]
- Foligne, B.; Nutten, S.; Grangette, C.; Dennin, V.; Goudercourt, D.; Poiret, S.; Dewulf, J.; Brassart, D.; Mercenier, A.; Pot, B. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J. Gastroenterol. 2007, 13, 236–243. [Google Scholar] [CrossRef] [Green Version]
- Kishino, S.; Takeuchi, M.; Park, S.-B.; Hirata, A.; Kitamura, N.; Kunisawa, J.; Hiroshi, K.; Iwamoto, R.; Isobe, Y.; Arita, M.; et al. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc. Natl. Acad. Sci. USA 2013, 110, 17808–17813. [Google Scholar] [CrossRef] [Green Version]
- Maragkoudakis, P.A.; Chingwaru, W.; Gradisnik, L.; Tsakalidou, E.; Cencic, A. Lactic acid bacteria efficiently protect human and animal intestinal epithelial and immune cells from enteric virus infection. Int. J. Food Microbiol. 2010, 141, S91–S97. [Google Scholar] [CrossRef]
- Olofsson, T.C.; Vásquez, A. Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Curr. Microbiol. 2008, 57, 356–363. [Google Scholar] [CrossRef]
- Vásquez, A.; Olofsson, T.C.; Sammataro, D. A scientific note on the lactic acid bacterial flora in honeybees in the USA—A comparison with bees from Sweden. Apidologie 2008, 40, 26–28. [Google Scholar] [CrossRef] [Green Version]
- Olofsson, T.C.; Butler, È.; Markowicz, P.; Lindholm, C.; Larsson, L.; Vásquez, A. Lactic acid bacterial symbionts in honeybees—an unknown key to honey’s antimicrobial and therapeutic activities. Int. Wound J. 2014, 13, 668–679. [Google Scholar] [CrossRef]
- Carina Audisio, M.; Torres, M.J.; Sabaté, D.C.; Ibarguren, C.; Apella, M.C. Properties of different lactic acid bacteria isolated from Apis mellifera L. bee-gut. Microbiol. Res. 2011, 166, 1–13. [Google Scholar] [CrossRef]
- McFrederick, Q.S.; Vuong, H.Q.; Rothman, J.A. Lactobacillus micheneri sp. nov., Lactobacillus timberlakei sp. nov. and Lactobacillus quenuiae sp. nov., lactic acid bacteria isolated from wild bees and flowers. Int. J. Syst. Evol. Microbiol. 2018, 68, 1879–1884. [Google Scholar] [CrossRef]
- Janashia, I.; Alaux, C. Specific immune stimulation by endogenous bacteria in honey bees (Hymenoptera: Apidae). J. Econ. Entomol. 2016, 109, 1474–1477. [Google Scholar] [CrossRef]
- Rokop, Z.P.; Horton, M.A.; Newton, I.L.G. Interactions between cooccurring lactic acid bacteria in honey bee hives. Appl. Environ. Microbiol. 2015, 81, 7261–7270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, K.E.; Sheehan, T.H.; Mott, B.M.; Maes, P.; Snyder, L.; Schwan, M.R.; Walton, A.; Jones, B.M.; Corby-Harris, V. Microbial Ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS ONE 2013, 8, e83125. [Google Scholar] [CrossRef] [Green Version]
- Bulgasem, B.Y.; Lani, M.N.; Hassan, Z.; Yusoff, W.M.W.; Fnaish, S.G. Antifungal activity of lactic acid bacteria strains isolated from natural honey against pathogenic candida species. Mycobiology 2016, 44, 302–309. [Google Scholar] [CrossRef] [Green Version]
- Aween, M.M.; Hassan, Z.; Muhialdin, B.J.; Eljamel, Y.A.; Al-Mabrok, A.S.W.; Lani, M.N. Antibacterial activity of Lactobacillus acidophilus strains isolated from honey marketed in Malaysia against selected multiple antibiotic resistant (MAR) gram-positive bacteria. J. Food Sci. 2012, 77, M364–M371. [Google Scholar] [CrossRef]
- Asama, T.; Arima, T.H.; Gomi, T.; Keishi, T.; Tani, H.; Kimura, Y.; Tatefuji, T.; Hashimoto, K. Lactobacillus kunkeei YB38 from honeybee products enhances IgA production in healthy adults. J. Appl. Microbiol. 2015, 119, 818–826. [Google Scholar] [CrossRef]
- Libonatti, C.; Agüeria, D.; García, C.; Basualdo, M. Weissella paramesenteroides encapsulation and its application in the use of fish waste. Revista Argentina de Microbiología 2018, 52, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Neveling, D.P.; Endo, A.; Dicks, L.M.T. Fructophilic Lactobacillus kunkeei and Lactobacillus brevis isolated from fresh flowers, bees and bee-hives. Curr. Microbiol. 2012, 65, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, J.; Ström, K.; Roos, S.; Sjögren, J.; Schnürer, J. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol. Lett. 2003, 219, 129–135. [Google Scholar] [CrossRef] [Green Version]
- Ruiz Rodríguez, L.G.; Mohamed, F.; Bleckwedel, J.; Medina, R.; De Vuyst, L.; Hebert, E.M.; Mozzi, F. Diversity and functional properties of lactic acid bacteria isolated from wild fruits and flowers present in northern Argentina. Front. Microbiol. 2019, 10, 1091. [Google Scholar] [CrossRef]
- Endo, A.; Futagawa-Endo, Y.; Sakamoto, M.; Kitahara, M.; Dicks, L.M.T. Lactobacillus florum sp. nov., a fructophilic species isolated from flowers. Int. J. Syst. Evol. Microbiol. 2010, 60, 2478–2482. [Google Scholar] [CrossRef]
- Ushio, M.; Yamasaki, E.; Takasu, H.; Nagano, A.J.; Fujinaga, S.; Honjo, M.N.; Ikemoto, M.; Sakai, S.; Kudoh, H. Microbial communities on flower surfaces act as signatures of pollinator visitation. Sci. Rep. 2015, 5, 8695. [Google Scholar] [CrossRef]
- Martinson, V.G.; Danforth, B.N.; Minckley, R.L.; Rueppell, O.; Tingek, S.; Moran, N.A. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 2010, 20, 619–628. [Google Scholar] [CrossRef]
- Wu, M.; Sugimura, Y.; Iwata, K.; Takaya, N.; Takamatsu, D.; Kobayashi, M.; Taylor, D.; Kimura, K.; Yoshiyama, M. Inhibitory effect of gut bacteria from the Japanese honey bee, Apis cerana japonica, against Melissococcus plutonius, the causal agent of European foulbrood disease. J. Insect. Sci. 2014, 14, 129. [Google Scholar] [CrossRef]
- Vahedi-Shahandashti, R.; Kasra-Kermanshahi, R.; Shokouhfard, M.; Ghadam, P.; Feizabadi, M.M.; Teimourian, S. Antagonistic activities of some probiotic lactobacilli culture supernatant on Serratia marcescens swarming motility and antibiotic resistance. Iran J. Microbiol. 2017, 9, 348–355. [Google Scholar]
- Arredondo, D.; Castelli, L.; Porrini, M.P.; Garrido, P.M.; Eguaras, M.J.; Zunino, P.; Antúnez, K. Lactobacillus kunkeei strains decreased the infection by honey bee pathogens Paenibacillus larvae and Nosema ceranae. Beneficial Microbes 2018, 9, 279–290. [Google Scholar] [CrossRef]
- Fünfhaus, A.; Ebeling, J.; Genersch., E. Bacterial pathogens of bees. Curr. Opin. Insect. Sci. 2018, 26, 89–96. [Google Scholar] [CrossRef]
- Nowak, I.; Nowak, A.; Leska, A. American foulbrood, as an infectious disease of honey bees—Selected legal and environmental aspects. Studies in Law and Economics 2020, 115, 87–108. [Google Scholar] [CrossRef]
- Peghaire, E.; Moné, A.; Delbac, F.; Debroas, D.; Chaucheyras-Durand, F.; El Alaoui, H. A Pediococcus strain to rescue honeybees by decreasing Nosema ceranae-and pesticide-induced adverse effects. Pestic. Biochem. Physiol. 2020, 163, 138–146. [Google Scholar] [CrossRef]
- Tejerina, M.R.; Cabana, M.J.; Benitez-Ahrendts, M.R. Strains of Lactobacillus spp. reduce chalkbrood in Apis mellifera. J. Invertebr. Pathol. 2020, 178, 107521. [Google Scholar] [CrossRef]
- Kačániová, M.; Terentjeva, M.; Žiarovská, J.; Kowalczewski, P.Ł. In vitro antagonistic effect of gut bacteriota isolated from indigenous honey bees and essential oils against Paenibacillus larvae. Int. J. Mol. Sci. 2020, 21, 6736. [Google Scholar] [CrossRef]
- Stephan, J.G.; Lamei, S.; Pettis, J.S.; Riesbeck, K.; de Miranda, J.R.; Forsgren, E. Honeybee-specific lactic acid bacterium supplements have no effect on American foulbrood-infected honeybee colonies. Appl. Environ. Microbiol. 2019, 17, 85–e00606. [Google Scholar] [CrossRef] [Green Version]
- Lamei, S.; Stephan, J.G.; Nilson, B.; Sieuwerts, S.; Riesbeck, K.; de Miranda, J.R.; Forsgren, E. Feeding honeybee colonies with honeybee-specific lactic acid bacteria (Hbs-LAB) does not affect colony-level Hbs-LAB composition or Paenibacillus larvae spore levels, although American foulbrood affected colonies harbor a more diverse Hbs-LAB community. Microb. Ecol. 2020, 79, 743–755. [Google Scholar] [CrossRef] [Green Version]
- Daisley, B.A.; Pitek, A.P.; Chmiel, J.A.; Al, K.F.; Chernyshova, A.M.; Faragalla, K.M.; Burton, J.P.; Thompson, G.J.; Reid, G. Novel probiotic approach to counter Paenibacillus larvae infection in honey bees. ISME J. 2020, 14, 476–491. [Google Scholar] [CrossRef] [Green Version]
- Ramos, O.Y.; Basualdo, M.; Libonatti, C.; Vega, M.F. Current status and application of lactic acid bacteria in animal production systems with a focus on bacteria from honey bee colonies. J. Appl. Microbiol. 2020, 128, 1248–1260. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Marenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nature Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Zommiti, M.; Feuilloley, M.G.J.; Connil, N. Update of Probiotics in Human World: A Nonstop Source of Benefactions till the End of Time. Microorganisms 2020, 8, 1907. [Google Scholar] [CrossRef]
- Nowak, A.; Paliwoda, A.; Błasiak, J. Anti-proliferative, pro-apoptotic and anti-oxidative activity of Lactobacillus and Bifidobacterium strains: A Review of mechanisms and therapeutic perspectives. Crit. Rev. Food Sci. Nutr. 2019, 59, 3456–3467. [Google Scholar] [CrossRef]
- Zhang, M.; Ming, Y.; Guo, H.; Zhu, Y.; Yang, T.; Chen, S.; He, L.; Ao, X.; Liu, A.; Zhou, K.; et al. Screening of lactic acid bacteria for their capacity to bind cypermethrin in vitro and the binding characteristics and its application. Food Chem. 2021, 347, 715–721. [Google Scholar] [CrossRef]
- Shi, Y.H.; Xiao, J.J.; Liu, Y.Y.; Deng, Y.J.; Feng, W.Z.; Wei, D.; Liao, M.; Cao, H.Q. Gut microbiota influence on oral bioaccessibility and intestinal transport of pesticides in Chaenomeles speciosa. Food Chem. 2021, 339, 127985. [Google Scholar] [CrossRef]
- Kumral, A.; Kumral, N.A.; Gurbuz, O. Chlorpyrifos and deltamethrin degradation potentials of two Lactobacillus plantarum (Orla-Jensen, 1919) (Lactobacillales: Lactobacillaceae) strains. Turkiye Entomoloji Dergisi 2020, 44, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Pinto, G.D.A.; Castro, I.M.; Miguel, M.A.L.; Koblitz, M.G.B. Lactic acid bacteria—promising technology for organophosphate degradation in food: A pilot study. LWT 2019, 110, 353–359. [Google Scholar] [CrossRef]
- Duan, J.; Cheng, Z.; Bi, J.; Xu, Y. Residue behavior of organochlorine pesticides during the production process of yogurt and cheese. Food Chem. 2018, 15, 119–124. [Google Scholar] [CrossRef]
- Li, C.; Ma, Y.; Mi, Z.; Huo, R.; Zhou, T.; Hai, H.; Kwok, L.Y.; Sun, Z.; Chen, Y.; Zhang, H. Screening for Lactobacillus plantarum Strains That Possess Organophosphorus Pesticide-Degrading Activity and Metabolomic Analysis of Phorate Degradation. Frot. Microbiol. 2018, 9, 2048. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.S.; Wu, T.H.; Yang, Y.; Zhu, C.L.; Ding, C.L.; Dai, C.C. Binding and detoxification of chlorpyrifos by lactic acid bacteria on rice straw silage fermentation. J. Environ. Sci. Health B 2016, 51, 316–325. [Google Scholar] [CrossRef]
- Trinder, M.; McDowell, T.W.; Daisley, B.A.; Ali, S.N.; Leong, H.S.; Sumarah, M.W.; Reid, G. Probiotic Lactobacillus rhamnosus Reduces Organophosphate Pesticide Absorption and Toxicity to Drosophila melanogaster. Appl. Environ. Microbiol. 2016, 82, 6204–6213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagherpour Shamloo, H.; Golkari, S.; Faghfoori, Z.; Movassaghpour, A.; Lotfi, H.; Barzegari, A.; Yari Khosroushahi, A. Lactobacillus casei decreases organophosphorus pesticide diazinon cytotoxicity in human HUVEC cell line. Adv. Pharm. Bull. 2016, 6, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Bouhafs, L.; Moudilou, E.N.; Exbrayat, J.M.; Lahouel, M.; Idoui, T. Protective effects of probiotic Lactobacillus plantarum BJ0021 on liver and kidney oxidative stress and apoptosis induced by endosulfan in pregnant rats. Ren. Fail. 2015, 37, 1370–1378. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; Xu, D.; Liu, J.Q.; Zhao, X.H. Enhanced degradation of five organophosphorus pesticides in skimmed milk by lactic acid bacteria and its potential relationship with phosphatase production. Food Chem. 2014, 164, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Dorđević, T.M.; Siler-Marinković, S.S.; Durović-Pejčev, R.D.; Dimitrijević-Branković, S.I.; Gajić Umiljendić, J.S. Dissipation of pirimiphos-methyl during wheat fermentation by Lactobacillus plantarum. Lett. Appl. Microbiol. 2013, 57, 412–419. [Google Scholar] [CrossRef]
- Harishankar, M.K.; Sasikala, C.; Ramya, M. Efficiency of the intestinal bacteria in the degradation of the toxic pesticide, chlorpyrifos. 3 Biotech 2013, 3, 137–142. [Google Scholar] [CrossRef] [Green Version]
- Islam, S.M.; Math, R.K.; Cho, K.M.; Lim, W.J.; Hong, S.Y.; Kim, J.M.; Yun, M.G.; Cho, J.J.; Yun, H.D. Organophosphorus hydrolase (OpdB) of Lactobacillus brevis WCP902 from kimchi is able to degrade organophosphorus pesticides. J. Agric. Food Chem. 2010, 12, 5380–5386. [Google Scholar] [CrossRef]
- Cho, K.M.; Math, R.K.; Islam, S.M.; Lim, W.J.; Hong, S.Y.; Kim, J.M.; Yun, M.G.; Cho, J.J.; Yun, H.D. Biodegradation of chlorpyrifos by lactic acid bacteria during kimchi fermentation. J. Agric. Food Chem. 2009, 57, 1882–1889. [Google Scholar] [CrossRef]
- Ptaszyńska, A.A.; Małek, W.; Borsuk, G.; Grzęda, M.; Wicha, M.; Pachla, A. Bacterial Strains of the Lactobacillus and Fructobacillus Genera, Isolated from Alimentary Tract of Honeybees to Be Applied for Fighting and Prevention of Bees’ Diseases and the Probiotic Preparations Based on Such Bacteria Strains. Application for Patent No. P.423363, 2017. The Patent Office of the Republic of Poland. Available online: https://ewyszukiwarka.pue.uprp.gov.pl (accessed on 21 March 2021).
- Maruščáková, I.C.; Schusterová, P.; Bielik, B.; Toporčák, J.; Bíliková, K.; Mudroňová, D. Effect of application of probiotic pollen suspension on immune response and gut microbiota of honey bees (Apis mellifera). Probiotics Antimicrob. Proteins 2020, 12, 929–936. [Google Scholar] [CrossRef]
- Tlak Gajger, I.; Vlainić, J.; Šoštarić, P.; Prešern, J.; Bubnič, J.; Smodiš Škerl, M.I. Effects on Some Therapeutical, Biochemical, and Immunological Parameters of Honey Bee (Apis mellifera) Exposed to Probiotic Treatments, in Field and Laboratory Conditions. Insects 2020, 11, 638. [Google Scholar] [CrossRef]
- Goblirsch, M.J.; Spivak, M.S.; Kurtti, T.J. A cell line resource derived from honey bee (Apis mellifera) embryonic tissues. PLoS ONE 2013, 8, e69831. [Google Scholar] [CrossRef] [Green Version]
- Genersch, E.; Gisder, S.; Hedtke, K.; Hunter, W.B.; Möckel, N.; Müller, U. Standard methods for cell cultures in Apis mellifera research. J. Apic. Res. 2013, 52, 1–8. [Google Scholar] [CrossRef] [Green Version]




| Preparation Name | Producer | Short Characteristics | Effects |
|---|---|---|---|
| Apiflora | Biowet, Poland | Lyophilized selected Lactobacillus strains; 1×108 CFU/vial; application in water or sugar syrup. Elaborated with Maria Curie-Skłodowska University in Lublin and University of Life Sciences in Lublin, Poland. | Colonization of honeybee gut. Antagonistic effect toward P. larvae and N. ceranae. Increase of honeybee survival rate. Available at: https://biowet.pl/en/produkty/apiflora-2/, accessed on 22 March 2021 |
| EM® PROBIOTIC FOR BEES | EMRO, Japan | Multiple species of lactic acid bacteria, yeast, and photosynthetic bacteria. No detailed information given. | Inhibition of nosemosis: reduction of spore counts in colonies; colonies’ strength increased. Positive physiological changes in probiotic-treated groups of adult bees [220]. |
| SuperDFM®-Honeybee | Strong Microbials, USA | Dried: L. acidophilus, Ent. faecium, B. bifidum, L. plantarum, Saccharomyces cerevisiae, Bacillus subtilis, Bacillus licheniformis, Bacillus pumilus fermentation products; dried B. subtilis fermentation extract. Total min. LAB count: 1.5 billion CFU/g. Total min. yeast count: 1 billion CFU/g. | Digestion and nutrient absorption improvement, gut health promotion, renewal of the microbes. Available at: https://www.strongmicrobials.com/honeybee, accessed on 22 March 2021 |
| SuperDFM® +P801™ | Strong Microbials, USA | Composition as in the case of SuperDFM®-Honeybee plus P. acidilactici P801 fermentation product. Total min. LAB count: 2 billion CFU/g. | Strengthen and stimulate the immune system, aiding optimal nutrient absorption, better survivorship to honeybees exposed to pesticides. Available at: https://www.strongmicrobials.com/superdfm-p801, accessed on 22 March 2021 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowak, A.; Szczuka, D.; Górczyńska, A.; Motyl, I.; Kręgiel, D. Characterization of Apis mellifera Gastrointestinal Microbiota and Lactic Acid Bacteria for Honeybee Protection—A Review. Cells 2021, 10, 701. https://doi.org/10.3390/cells10030701
Nowak A, Szczuka D, Górczyńska A, Motyl I, Kręgiel D. Characterization of Apis mellifera Gastrointestinal Microbiota and Lactic Acid Bacteria for Honeybee Protection—A Review. Cells. 2021; 10(3):701. https://doi.org/10.3390/cells10030701
Chicago/Turabian StyleNowak, Adriana, Daria Szczuka, Anna Górczyńska, Ilona Motyl, and Dorota Kręgiel. 2021. "Characterization of Apis mellifera Gastrointestinal Microbiota and Lactic Acid Bacteria for Honeybee Protection—A Review" Cells 10, no. 3: 701. https://doi.org/10.3390/cells10030701