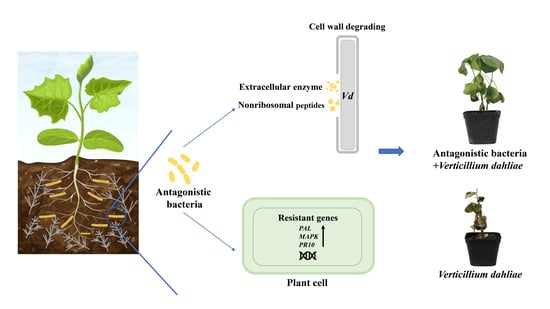
Potential Antagonistic Bacteria against Verticillium dahliae Isolated from Artificially Infested Nursery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil Samples Collection
2.2. V. dahliae Strain
2.3. Isolation of Potential Antagonistic Bacteria
2.4. In Vitro Antagonism Test
2.5. Molecular Identification of Antagonistic Bacteria and Phylogenetic Correlation
2.6. Extracellular Enzyme Activity Assays
2.7. Lipopeptides Detection
2.8. The Expression Profile Analysis of Cotton Resistance Genes
2.9. Disease Index
2.10. Fungal Biomass
2.11. Statistical Analysis
3. Results
3.1. Isolation and Phylogenetic Relationship Analysis of Antagonistic Bacteria against V. dahliae
3.2. In Vitro Antagonism Test
3.3. Extracellular Enzyme Activity
3.4. Lipopeptides Production
3.5. Cotton Resistant Genes Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fradin, E.F.; Thomma, B.P. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant. Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef] [Green Version]
- Huisman, O. Interrelations of root growth dynamics to epidemiology of root-invading fungi. Annu. Rev. Phytopathol. 1982, 20, 303–327. [Google Scholar] [CrossRef]
- Tsror, L.; Levin, A.G. Vegetative compatibility and pathogenicity of Verticillium dahliae Kleb. Isolates from olive in Israel. J. Phytopathol. 2003, 151, 451–455. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, C.; Wu, S.; Zhang, X.; Tang, J.; Jian, G.; Jiao, G.; Li, F.; Chu, C. Significant improvement of cotton Verticillium wilt resistance by manipulating the expression of gastrodia antifungal proteins. Mol. Plant. 2016, 9, 1436–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heale, J.B.; Isaac, I. Environmental factors in the production of dark resting structures in Verticillium alboatrum, V. dahliae and V. tricorpus. Trans. Br. Mycol. Soc. 1965, 48, 39–50. [Google Scholar] [CrossRef]
- Mol, L.; Riessen, H.W.V. Effect of plant roots on the germination of microsclerotia of Verticillum dahliae. Eur. J. Plant. Pathol. 1995, 101, 673–678. [Google Scholar] [CrossRef]
- Hu, X.; Bai, Y.; Chen, T.; Hu, D.; Yang, J.; Xu, X. An optimized method for in vitro production of Verticillium dahliae microsclerotia. Eur. J. Plant. Pathol. 2013, 136, 225–229. [Google Scholar] [CrossRef]
- Farley, J.D.; Wilhelm, S.; Snyder, W.C. Repeated germination and sporulation of microsclerotia of Verticillium albo-atrum in soil. Phytopathology 1971, 61, 260–264. [Google Scholar] [CrossRef]
- Reusche, M.; Truskina, J.; Thole, K.; Nagel, L.; Rindfleisch, S.; Tran, V.T.; Braus-Stromeyer, S.A.; Braus, G.H.; Teichmann, T.; Lipka, V. Infections with the vascular pathogens Verticillium longisporum and Verticillium dahliae induce distinct disease symptoms and differentially affect drought stress tolerance of Arabidopsis thaliana. Environ. Exp. Bot. 2014, 108, 23–37. [Google Scholar] [CrossRef]
- Heinz, R.; Lee, S.W.; Saparno, A.; Nazar, R.N.; Robb, J. Cyclical systemic colonization in Verticillium-infected tomato. Physiol. Mol. Plant. Pathol. 1998, 52, 385–396. [Google Scholar] [CrossRef]
- Skadow, K. Weeds as host-plants for Verticillium albo-atrum Rke. et berth. Including Verticillium dahliae Kleb. Zent. Bakteriol. Parasitenkd. Infekt. Hyg. 1966, 120, 49–59. [Google Scholar]
- Powelson, M.L.; Rowe, R.C. Biology and management of early dying of potatoes. Annu. Rev. Phytopathol. 1993, 31, 111–126. [Google Scholar] [CrossRef]
- Tohidfar, M.; Mohammadi, M.; Ghareyazie, B. Agrobacterium-mediated transformation of cotton (Gossypium hirsutum) using a heterologous bean chitinase gene. Plant. Cell Tissue Organ. Cult. 2005, 83, 83–96. [Google Scholar] [CrossRef]
- Davis, J. Effects of green manures on Verticillium wilt of potato. Phytopathology 1996, 86, 444–453. [Google Scholar] [CrossRef]
- Tohidfar, M.; Hossaini, R.; Bashir, N.S.; Meisam, T. Enhanced resistance to Verticillium dahliae in transgenic cotton expressing an endochitinase gene from Phaseolus vulgaris. Czech. J. Genet. Plant. Breed. 2012, 48, 33–41. [Google Scholar] [CrossRef] [Green Version]
- Arbogast, M.; Powelson, M.L.; Cappaert, M.R.; Watrud, L.S. Response of six potato cultivars to amount of applied water and Verticillium dahliae. Phytopathology 1999, 89, 782–788. [Google Scholar] [CrossRef] [Green Version]
- Mendgen, K.; Hahn, M. Plant infection and the establishment of fungal biotrophy. Trends Plant Sci. 2002, 7, 352–356. [Google Scholar] [CrossRef] [Green Version]
- Longnecker, M.P.; Rogan, W.J.; Lucier, G. The human health effects of DTT (dichlorodiphenyltrichloroethane) and PCBs (polychlorinated biphenyls) and an overview of organochlorines in public health. Annu. Rev. Public Health 1997, 18, 211–244. [Google Scholar] [CrossRef] [Green Version]
- Manuel, A.-E.; Eugenio, L.-P.; Elena, M.-C.; Simal-Gándara, J.; Mejuto, J.-C.; García-Río, L. The mobility and degradation of pesticides in soils and the pollution of groundwater resources. Agric. Ecosyst. Environ. 2008, 123, 247–260. [Google Scholar]
- Chang, S.H.; Kwack, M.S.; Kim, Y.S.; Lee, J.Y.; Kim, K.D. A rapid radicle assay for prescreening antagonistic bacteria against Phytophthora capsici on pepper. Mycobiology 2001, 29, 218–223. [Google Scholar] [CrossRef]
- Cao, H.; He, S.; Wei, R.; Diong, M.; Lu, L. Bacillus amyloliquefaciens G1: A potential antagonistic bacterium against eel-pathogenic Aeromonas hydrophila. Evid. Based Complementary Altern. Med. Ecam. 2011, 2011, 824104–824110. [Google Scholar] [CrossRef] [Green Version]
- Karimi, S.; Rashidian, E.; Birjandi, M.; Mahmoodnia, L. Antagonistic effect of isolated probiotic bacteria from natural sources against intestinal Escherichia coli pathotypes. Electron. Physician 2018, 10, 6534–6539. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.C.; Korkor, N.L.; Xiao, R.; Pu, Q.; Hu, M.; Zhang, S.S.; Kong, D.D.; Zeng, G.; Hu, X.F. Antagonistic activity of combined bacteria strains against southern blight pathogen of Dendrobium officinale. Biol. Control. 2020, 151, 104291–104301. [Google Scholar] [CrossRef]
- Kunova, A.; Bonaldi, M.; Saracchi, M.; Pizzatti, C.; Chen, X.; Cortesi, P. Selection of Streptomyces against soil borne fungal pathogens by a standardized dual culture assay and evaluation of their effects on seed germination and plant growth. BMC Microbiol. 2016, 16, 272–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faria, D.C.; Dias, A.C.F.; Melo, I.S.; Carvalho Costa, F.E. Endophytic bacteria isolated from orchid and their potential to promote plant growth. World J. Microbiol. Biotechnol. 2013, 29, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Kurt, S.; Dervis, S.; Sahinler, S. Sensitivity of Verticillium dahliae to prochloraz and prochloraz–manganese complex and control of Verticillium wilt of cotton in the field. Crop. Protect. 2003, 22, 51–55. [Google Scholar] [CrossRef]
- Tehrani, A.S.; Disfani, F.A.; Hedjaroud, G.A.; Mohammadi, M. Antagonistic effects of several bacteria on Verticillium dahliae the causal agent of cotton wilt. Meded. Rijksuniv. Gent Fak. Landbouwkd. Toegep. Biol. Wet. 2001, 66, 95–101. [Google Scholar] [PubMed]
- Tjamos, E.C.; Tsitsigiannis, D.I.; Tjamos, S.E.; Antoniou, P.P.; Katinakis, P. Selection and screening of endorhizosphere bacteria from solarized soils as biocontrol agents against Verticillium dahliae of solanaceous hosts. Eur. J. Plant. Pathol. 2004, 110, 35–44. [Google Scholar] [CrossRef]
- Erdogan, O.; Benlioglu, K. Biological control of Verticillium wilt on cotton by the use of fluorescent Pseudomonas spp. Under field conditions. Biol. Control 2010, 53, 39–45. [Google Scholar] [CrossRef]
- Li, S.; Zhang, N.; Zhang, Z.; Luo, J.; Shen, B.; Zhang, R.; Shen, Q. Antagonist Bacillus subtilis HJ5 controls Verticillium wilt of cotton by root colonization and biofilm formation. Biol. Fertil. Soils 2012, 49, 295–303. [Google Scholar] [CrossRef]
- Zhou, J.; Feng, Z.; Feng, H.; Li, Y.; Yuan, Y.; Li, Z.; Wei, F.; Shi, Y.; Zhao, L.; Sun, Z.; et al. Biocontrol effect and mechanism of cotton endophytic bacterium Bacillus cereus YUPP-10 against Verticillium wilt in Gossypium hirsutum. Sci. Agric. Sin. 2017, 50, 2717–2727. [Google Scholar]
- Safdarpour, F.; Khodakaramian, G. Assessment of antagonistic and plant growth promoting activities of tomato endophytic bacteria in challenging with Verticillium dahliae under in-vitro and in-vivo conditions. Biol. J. Microorg. 2019, 7, 77–90. [Google Scholar]
- Xue, L.; Xue, Q.H.; Chen, Q.; Lin, C.F.; Shen, G.H.; Zhao, J. Isolation and evaluation of rhizosphere actinomycetes with potential application for biocontrol of Verticillium wilt of cotton. Crop. Protect. 2013, 43, 231–240. [Google Scholar] [CrossRef]
- Li, Q.; Liao, S.; Wei, J.; Xing, D.; Xiao, Y.; Yang, Q. Isolation of Bacillus subtilis strain SEM-2 from silkworm excrement and characterization of its antagonistic effect against Fusarium spp. Can. J. Microbiol. 2020, 66, 401–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, D. 16s/23s rRNA sequencing. Nucleic Acid Tech. Bact. Syst. 1991, 115–175. [Google Scholar] [CrossRef]
- Wang, P.; Guo, Q.; Ma, Y.; Li, S.; Lu, X.; Zhang, X.; Ma, P. DegQ regulates the production of fengycins and biofilm formation of the biocontrol agent Bacillus subtilis NCD-2. Microbiol. Res. 2015, 178, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, K.; Wollherr, A.; Larsen, M.; Rachinger, M.; Liesegang, H.; Ehrenreich, A.; Meinhardt, F. Facilitation of direct conditional knockout of essential genes in Bacillus licheniformis DSM13 by comparative genetic analysis and manipulation of genetic competence. Appl. Environ. Microbiol. 2010, 76, 5046–5057. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, M. Isolation purification and characterization of glucanase enzyme from antagonistic fungus Trichoderma. Int. J. Sci. Eng. Res. 2014, 5, 646–649. [Google Scholar]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Biochem. 2009, 55, 611–622. [Google Scholar]
- Su, X.; Lu, G.; Li, X.; Rehman, L.; Liu, W.; Sun, G.; Guo, H.; Wang, G.; Cheng, H. Host-induced gene silencing of an adenylate kinase gene involved in fungal energy metabolism improves plant resistance to Verticillium dahliae. Biomolecules 2020, 10, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Sun, L.; Wassan, G.M.; He, X.; Shaban, M.; Zhang, L.; Zhu, L.; Zhang, X. GbSOBIR1 confers Verticillium wilt resistance by phosphorylating the transcriptional factor GBbHLH171 in Gossypium barbadense. Plant. Biotechnol. J. 2019, 17, 152–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, X.; Rehman, L.; Guo, H.; Li, X.; Cheng, H. The oligosaccharyl transferase subunit STT3 mediates fungal development and is required for virulence in Verticillium dahliae. Curr. Genet. 2018, 64, 235–246. [Google Scholar] [CrossRef]
- Li, X.; Su, X.; Lu, G.; Sun, G.; Zhang, Z.; Guo, H.; Guo, N.; Cheng, H. VdOGDH is involved in energy metabolism and required for virulence of Verticillium dahliae. Curr. Genet. 2019, 66, 345–359. [Google Scholar] [CrossRef]
- Tzima, A.K.; Paplomatas, E.J.; Tsitsigiannis, D.I.; Kang, S. The G protein beta subunit controls virulence and multiple growth- and development-related traits in Verticillium dahliae. Fungal Genet. Biol. 2012, 49, 271–283. [Google Scholar] [CrossRef]
- Deravel, J.; Lemiere, S.; Coutte, F.; Krier, F.; Van Hese, N.; Bechet, M.; Sourdeau, N.; Hofte, M.; Lepretre, A.; Jacques, P. Mycosubtilin and surfactin are efficient, low ecotoxicity molecules for the biocontrol of lettuce downy mildew. Appl. Microbiol. Biotechnol. 2014, 98, 6255–6264. [Google Scholar] [CrossRef]
- Govindasamy, V.; Senthilkumar, M.; Magheshwaran, V.; Kumar, U.; Bose, P.; Sharma, V.; Annapurna, K. Bacillus and Paenibacillus spp.: Potential PGPR for sustainable agriculture. In Plant Growth and Health Promoting Bacteria; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 333–364. [Google Scholar]
- van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castano-Espriu, L.; Chang, C.; Clark, T.N.; et al. The natural products atlas: An open access knowledge base for microbial natural products discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef] [Green Version]
- Flissi, A.; Ricart, E.; Campart, C.; Chevalier, M.; Dufresne, Y.; Michalik, J.; Jacques, P.; Flahaut, C.; Lisacek, F.; Leclere, V.; et al. Norine: Update of the nonribosomal peptide resource. Nucleic Acids Res. 2020, 48, D465–D469. [Google Scholar]
- Gray, E.J.; Smith, D.L. Intracellular and extracellular PGPR: Commonalities and distinctions in the plant-bacterium signaling processes. Soil Biol. Biochem. 2005, 37, 395–412. [Google Scholar] [CrossRef]
- Dobbelaere, S.; Vanderleyden, J.; Okon, Y. Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit. Rev. Plant. Sci. 2002, 22, 107–149. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podile, A.R.; Kishore, G.K. Plant growth-promoting rhizobacteria. In Plant-Associated Bacteria; Gnanamanickam, S.S., Ed.; Springer: Dordrecht, The Netherlands, 2006; pp. 195–230. [Google Scholar]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, G.; Li, D.; Li, S.; Hong, Q. Identification and evaluation of strain B37 of Bacillus subtilis antagonistic to sapstain fungi on poplar wood. Sci. World J. 2014, 2014, 149342–149351. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Hu, M.; Xue, Y.; Chen, X.; Lu, G.; Zhang, L.; Zhou, J. Screening, identification and efficacy evaluation of antagonistic bacteria for biocontrol of soft rot disease caused by Dickeya zeae. Microorganisms 2020, 8, 697. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Nasim, F.U.; Ruhi, R.; Murad, H.; Ejaz, S.; Choudhary, M.S.; Mustafa, G.; Ashraf, M.; Rehman, J. Isolation and characteristics of biotechnologically important antagonistic thermophilic bacteria from rhizosphere of Haloxylon salicornicum. Pol. J. Microbiol. 2018, 67, 49–58. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, Z.; Wu, J.; Chen, L.; Dong, W. Isolated Bacillus subtilis strain 330-2 and its antagonistic genes identified by the removing PCR. Sci. Rep. 2017, 7, 1777–1789. [Google Scholar] [CrossRef] [Green Version]
- Benizri, E.; Baudoin, E.; Guckert, A. Root colonization by inoculated plant growth-promoting rhizobacteria. Biocontrol Sci. Technol. 2010, 11, 557–574. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Herrera-Estrella, A.; Seidl-Seiboth, V.; Martinez, D.A.; Druzhinina, I.S.; Thon, M.; Zeilinger, S.; Casas-Flores, S.; Horwitz, B.A.; Mukherjee, P.K.; et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011, 12, 40–54. [Google Scholar] [CrossRef] [Green Version]
- Cai, F.; Chen, W.; Wei, Z.; Pang, G.; Li, R.; Ran, W.; Shen, Q. Colonization of Trichoderma harzianum strain SQR-T037 on tomato roots and its relationship to plant growth, nutrient availability and soil microflora. Plant. Soil 2014, 388, 337–350. [Google Scholar] [CrossRef]
- Langner, T.; Gohre, V. Fungal chitinases: Function, regulation, and potential roles in plant/pathogen interactions. Curr. Genet. 2016, 62, 243–254. [Google Scholar] [CrossRef]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. Bioessays 2006, 28, 799–808. [Google Scholar] [CrossRef]
- Cabib, E.; Bowers, B.; Sburlati, A.; Silverman, S.J. Fungal cell wall synthesis: The construction of a biological structure. Microbiol. Sci. 1988, 5, 370–375. [Google Scholar]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LySM receptor molecules, CEBiP andOsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant. J. 2010, 64, 204–214. [Google Scholar] [CrossRef] [Green Version]
- Pusztahelyi, T. Chitin and chitin-related compounds in plant-fungal interactions. Mycology 2018, 9, 189–201. [Google Scholar] [CrossRef]
- Dubery, I.A.; Slater, V. Induced defence responses in cotton leaf disks by elicitors from Verticillium dahliae. Phytochemistry 1997, 44, 1429–1434. [Google Scholar] [CrossRef]
- Asaka, O.; Shoda, M. Biocontrol of Rhizoctonia solani damping-off of tomato with Bacillus subtilis RB14. Appl. Environ. Microbiol. 1996, 62, 4081–4085. [Google Scholar] [CrossRef] [Green Version]
- Kaspar, F.; Neubauer, P.; Gimpel, M. Bioactive secondary metabolites from Bacillus subtilis: A comprehensive review. J. Nat. Prod. 2019, 82, 2038–2053. [Google Scholar] [CrossRef] [PubMed]
- Shafi, J.; Tian, H.; Ji, M. Bacillus species as versatile weapons for plant pathogens: A review. Biotechnol. Biotechnol. Equip. 2017, 31, 446–459. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, D.K.; Johri, B.N. Interactions of Bacillus spp. And plants–with special reference to induced systemic resistance (ISR). Microbiol. Res. 2009, 164, 493–513. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, M.R.B.; Vilcinskas, A.; Rahnamaeian, M. Cooperative interaction of antimicrobial peptides with the interrelated immune pathways in plants. Mol. Plant. Pathol. 2016, 17, 464–471. [Google Scholar] [CrossRef] [Green Version]
- Jamil, S.; Copley, T.R.; Jabaji, S.H.; Rita, G. Antioxidant genes of plants and fungal pathogens are distinctly regulated during disease development in different Rhizoctonia solani pathosystems. PLoS ONE 2018, 13, e0192682. [Google Scholar]
- Chen, J.; Dou, k.; Gao, Y.D.; QIan, L.Y. Mechanism and application of Trichoderma spp. In biological control of corn diseases. Mycosystema 2014, 33, 1154–1167. [Google Scholar]
- Zhou, J.; Feng, Z.; Liu, S.; Wei, F.; Shi, Y.; Zhao, L.; Huang, W.; Zhou, Y.; Feng, H.; Zhu, H. CGTase, a novel antimicrobial protein from Bacillus cereus YUPP-10, suppresses Verticillium dahliae and mediates plant defence responses. Mol. Plant. Pathol. 2020, 22, 130–144. [Google Scholar] [CrossRef]
- Ningbo, W.; Mengjie, L.; Lihua, G.; Xiufen, Y.; Dewen, Q. A novel protein elicitor (PeBA1) from Bacillus amyloliquefaciens NC6 induces systemic resistance in tobacco. Int. J. Biol. Sci. 2016, 12, 757–767. [Google Scholar]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Agarwal, P.K. Pathogenesis related-10 proteins are small, structurally similar but with diverse role in stress signaling. Mol. Biol. Rep. 2014, 41, 599–611. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, X.; Wu, S.; Liu, L.; Lu, G.; Liu, H.; Jin, X.; Wang, Y.; Guo, H.; Wang, C.; Cheng, H. Potential Antagonistic Bacteria against Verticillium dahliae Isolated from Artificially Infested Nursery. Cells 2021, 10, 3588. https://doi.org/10.3390/cells10123588
Su X, Wu S, Liu L, Lu G, Liu H, Jin X, Wang Y, Guo H, Wang C, Cheng H. Potential Antagonistic Bacteria against Verticillium dahliae Isolated from Artificially Infested Nursery. Cells. 2021; 10(12):3588. https://doi.org/10.3390/cells10123588
Chicago/Turabian StyleSu, Xiaofeng, Siyuan Wu, Lu Liu, Guoqing Lu, Haiyang Liu, Xi Jin, Yi Wang, Huiming Guo, Chen Wang, and Hongmei Cheng. 2021. "Potential Antagonistic Bacteria against Verticillium dahliae Isolated from Artificially Infested Nursery" Cells 10, no. 12: 3588. https://doi.org/10.3390/cells10123588