Exosomes in Ageing and Motor Neurone Disease: Biogenesis, Uptake Mechanisms, Modifications in Disease and Uses in the Development of Biomarkers and Therapeutics
Abstract
:1. Introduction
2. Exosome Biogenesis and Secretion
2.1. ESCRT-Dependent Mechanism
2.2. ESCRT-Independent Mechanisms
2.2.1. Lipid-Mediated Biogenesis
2.2.2. Tetraspanin-Mediated Biogenesis
2.3. Release of Exosomes in the Extracellular Space
3. Heterogeneity in the Exosome Population
3.1. Exosome Membrane Composition
3.2. Exosome Lumen Content
3.2.1. Proteins
3.2.2. Nucleic Acid
3.3. Heterogenous Buoyant Properties
4. Exosomal Interaction with Recipient Cells and the Fate of Exosomes
4.1. Ligand–Receptor Interaction (Cell-Surface and Exosome-Surface Receptors)
4.2. Indirect Communication: Soluble Ligand Mediated Signalling
4.3. Fusion of Exosomes with the Plasma Membrane
4.4. Endocytosis
4.4.1. Clathrin-Mediated and Caveolin-Dependent Endocytosis
4.4.2. Macropinocytosis and Phagocytosis
4.4.3. Lipid-Raft-Mediated Endocytosis
4.5. Fate of Exosomes within Targeted Cells
5. Exosome-Induced Signaling in CNS; Role in Ageing and in the Neuromotor System
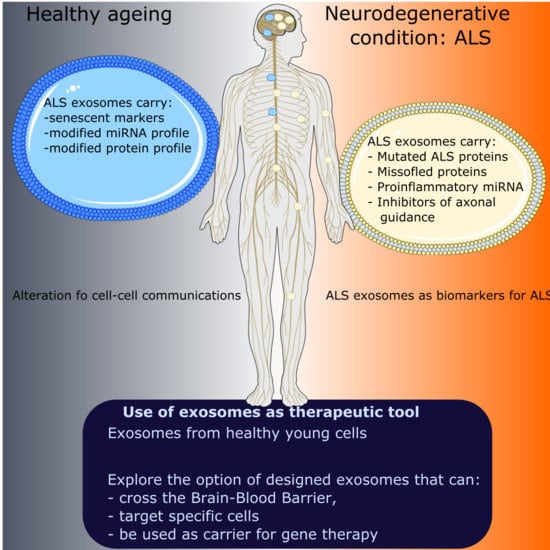
5.1. Impact of Ageing on Cell–Cell Communication
5.2. Secretion of sEVs by Senescent Cells
5.3. Proteins and miRNAs Associated with Senescence Contained within sEVs
5.4. sEV Therapeutics in Ageing
6. Exosomes in Neurodegenerative Disease Associated with Ageing: ALS
6.1. Detection of ALS Proteins in Exosomes: Potential Role of Exosomes in the Propagation of ALS
6.2. Potential Role of Exosomes to Modify Pathways in Recipient Cells in ALS
7. Use of Exosomes in Therapeutic Strategies for Neurodegenerative and Neuromuscular Conditions
7.1. Unmodified Exosomes as Therapeutics in Motor Neuron Disease
7.2. The Possibility to Use Modified Exosomes as Therapeutic Vehicles: Lessons from Other Neurodegenerative and Neuromuscular Conditions
8. Exosomes as Molecular Biomarkers for ALS
| Biofluid | sEV Isolation Technique | Exosome Parameters (Size, Exosome Marker). | Study Design | Exosome Origin & Analysis | Study Summary | Possible Biomarker | Ref |
|---|---|---|---|---|---|---|---|
| Plasma | Polymer-based precipitation and immunoaffinity purification using anti-CD-171 | 102 nm, CD63(+), TSG101(+), Calnexin (−) Neural Markers: L1CAM (+), NCAM 1 (+), MAPT (+), GRIA 1 (+), PLP 1 (+) | HC: 20 ALS: 20 Type: Not disclosed Age ≥ 18 yrs. old ALSFRS-R > 25 and FVC score ≥ 60%. | Neuron-derived exosomes Next generation sequencing (NGS) analysis, then downstream qPCR | 3 miRNAs downregulated and 5 miRNAs upregulated consistent and significant in ALS cohorts | miRNA fingerprinting for early ALS diagnosis | [286] |
| Plasma | Polymer-based precipitation and immunoaffinity purification using anti-CD-171. | 150 nm CD81(+), CD63(+) SNAP25 (+) Synaptophysin (+) [Neuron specific markers]. | HC: 5 ALS: 5 Type: Sporadic ALS Age and Sex matched. | Neuron-derived Exosomes Microarray analysis. | 30 differentially regulated miRNAs in ALS. miRNA upregulated (ALS): 13 miRNA downregulated (ALS): 17 | miRNAs within neuron-derived exosomes might be clinically advantageous in ALS diagnosis. | [283] |
| Plasma | Polymer-based precipitation and immunoprecipitation with biotinylated mouse anti-human glutamine aspartate transporter (ACSA-1) antibody. | 100 nm CD63 (+), Calnexin (−) | Recruitment: HC: 40 ALS: 39 For Study: HC: 12 ALS: 15 Type: Sporadic ALS (Bulbar onset:12; Limber onset: 28). ALSFRS-R: 39.83 ± 1.08 Age and Sex matched. | Astrocyte-derived Exosomes Enzyme-linked immunosorbent assay (ELISA) | Interleukin-6 (IL-6) levels increased in all ALS subgroups with no significant difference. Positive correlation between IL-6 levels and disease progression rate but not with total ALSFRS-R scores, diagnosis delay or patient age. For ALS < 12 months, Positive correlation between IL-6 levels in ADEs from ALS patients and rate of disease progression | IL-6 possible biomarker? Need for further studies and larger sample size. | [282] |
| Plasma | Heat Shock Protein- Vn96 synthetic peptide isolation followed by Centrifugation. | Nil parameters presented | HC: 12 ALS: 14 Type: Sporadic ALS (5M, 7F) Familial ALS (1M, 1F) ALSFRS-R: 26.23 ± 8.09 | droplet digital PCR-based miRNA quantification | 27 differentially regulated miRNAs in ALS. miR-15a-5p and miR-15a-5p/miR-181b-1-5p combination show diagnostic potential. miR-193a-5p distinguishes PALS with low and high ALSFRS-R scores. | miR-15a-5p and miR-193a-5p can be aid diagnosis and monitor ALS progression. | [281] |
| Serum | Polymer-based precipitation OR membrane affinity isolation | CD63 (+) | HC: 20 ALS: 10 Nil information on age matched or ALFRS score | Serum Exosomes Quantitative real-time PCR (qRT-PCR) | Downregulated expression of miR-27a-3p in ALS group that was statistically significant. | miR-27a3p as a reference for ALS diagnosis. | [284] |
| CSF | Sample concentration followed by Size Exclusion Chromatography and/or Ultracentrifugation. | 30–150 nm, CD81 (+), CD9 (+) | HC (iNPH): 3 ALS: 3 Type: Sporadic ALS Age and Sex matched. ALSFRS-R: 42.00 ± 1.00 | CSF-exosomes (exosome-enriched fractions from CSF) Proteomics | 334 proteins were identified including NIR (Novel INHAT Repressor) which was significantly increased in exosomes. | NIR as ALS biomarker and role in pathogenesis | [279] |
| CSF | Centrifugation | 186 nm ± 70.4 nm CD9 (+), CD81 (+) Flotillin-1 (+) | HC: 4 ALS: 4 Type: Sporadic ALS Age and Sex matched. ALSFRS-R: 41–45 Disease duration: 0.5–5 years | Neuronal-derived Exosomes Next-generation sequencing and qRT-PCR | 543 genes were significantly changed between HC and ALS groups. Genes upregulated (ALS):133 Genes downregulated (ALS): 410 | CUEDC2 (most increased exosomal mRNA in CSF from ALS group) | [280] |
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Nawaz, M.; Fatima, F. Extracellular vesicles, tunneling nanotubes, and cellular interplay: Synergies and missing links. Front. Mol. Biosci. 2017, 4, 50. [Google Scholar] [CrossRef] [Green Version]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Trams, E.G.; Lauter, C.J.; Salem, N.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fauré, J.; Lachenal, G.; Court, M.; Hirrlinger, J.; Chatellard-Causse, C.; Blot, B.; Grange, J.; Schoehn, G.; Goldberg, Y.; Boyer, V.; et al. Exosomes are released by cultured cortical neurones. Mol. Cell. Neurosci. 2006, 31, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Guescini, M.; Genedani, S.; Stocchi, V.; Agnati, L.F. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J. Neural Transm. 2010, 117, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Krämer-Albers, E.-M.; Bretz, N.; Tenzer, S.; Winterstein, C.; Möbius, W.; Berger, H.; Nave, K.-A.; Schild, H.; Trotter, J. Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: Trophic support for axons? Proteomics. Clin. Appl. 2007, 1, 1446–1461. [Google Scholar] [CrossRef] [PubMed]
- Potolicchio, I.; Carven, G.J.; Xu, X.; Stipp, C.; Riese, R.J.; Stern, L.J.; Santambrogio, L. Proteomic Analysis of Microglia-Derived Exosomes: Metabolic Role of the Aminopeptidase CD13 in Neuropeptide Catabolism. J. Immunol. 2005, 175, 2237–2243. [Google Scholar] [CrossRef] [Green Version]
- Frühbeis, C.; Fröhlich, D.; Kuo, W.P.; Amphornrat, J.; Thilemann, S.; Saab, A.S.; Kirchhoff, F.; Möbius, W.; Goebbels, S.; Nave, K.A.; et al. Neurotransmitter-Triggered Transfer of Exosomes Mediates Oligodendrocyte-Neuron Communication. PLoS Biol. 2013, 11, e1001604. [Google Scholar] [CrossRef] [Green Version]
- Fröhlich, D.; Kuo, W.P.; Frühbeis, C.; Sun, J.J.; Zehendner, C.M.; Luhmann, H.J.; Pinto, S.; Toedling, J.; Trotter, J.; Krämer-Albers, E.M. Multifaceted effects of oligodendroglial exosomes on neurons: Impact on neuronal firing rate, signal transduction and gene regulation. R. Soc. 2014, 369, 20130510. [Google Scholar] [CrossRef]
- Abbott, N.J.; Friedman, A. Overview and introduction: The blood-brain barrier in health and disease. Epilepsia 2012, 53 (Suppl. 6), 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zagrean, A.-M.; Hermann, D.M.; Opris, I.; Zagrean, L.; Popa-Wagner, A. Multicellular Crosstalk Between Exosomes and the Neurovascular Unit After Cerebral Ischemia. Therapeutic Implications. Front. Neurosci. 2018, 12, 811. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef] [Green Version]
- Emmanouilidou, E.; Melachroinou, K.; Roumeliotis, T.; Garbis, S.D.; Ntzouni, M.; Margaritis, L.H.; Stefanis, L.; Vekrellis, K. Cell-Produced -Synuclein Is Secreted in a Calcium-Dependent Manner by Exosomes and Impacts Neuronal Survival. J. Neurosci. 2010, 30, 6838–6851. [Google Scholar] [CrossRef] [Green Version]
- Silverman, J.M.; Christy, D.; Shyu, C.C.; Moon, K.-M.; Fernando, S.; Gidden, Z.; Cowan, C.M.; Ban, Y.; Stacey, R.G.; Grad, L.I.; et al. CNS-derived extracellular vesicles from superoxide dismutase 1 (SOD1)G93A ALS mice originate from astrocytes and neurons and carry misfolded SOD1. J. Biol. Chem. 2019, 294, 3744–3759. [Google Scholar] [CrossRef] [Green Version]
- Brites, D.; Vaz, A.R. Microglia centered pathogenesis in ALS: Insights in cell interconnectivity. Front. Cell. Neurosci. 2014, 8, 117. [Google Scholar] [CrossRef]
- Vijayakumar, U.G.; Milla, V.; Cynthia Stafford, M.Y.; Bjourson, A.J.; Duddy, W.; Duguez, S.M.-R. A Systematic Review of Suggested Molecular Strata, Biomarkers and Their Tissue Sources in ALS. Front. Neurol. 2019, 10, 400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Boussac, M.; Véron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [Green Version]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular Endosome Biogenesis in the Absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Babst, M. A Protein’s Final ESCRT. Traffic 2005, 6, 2–9. [Google Scholar] [CrossRef]
- Williams, R.L.; Urbé, S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007, 8, 355–368. [Google Scholar] [CrossRef]
- Kalra, H.; Drummen, G.; Mathivanan, S.; Kalra, H.; Drummen, G.P.C.; Mathivanan, S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [Green Version]
- Hanson, P.I.; Cashikar, A. Multivesicular Body Morphogenesis. Annu. Rev. Cell Dev. Biol. 2012, 28, 337–362. [Google Scholar] [CrossRef]
- Babst, M.; Katzmann, D.J.; Estepa-Sabal, E.J.; Meerloo, T.; Emr, S.D. ESCRT-III: An endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 2002, 3, 371–382. [Google Scholar] [CrossRef] [Green Version]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Ghossoub, R.; Lembo, F.; Rubio, A.; Gaillard, C.B.; Bouchet, J.; Vitale, N.; Slavík, J.; Machala, M.; Zimmermann, P. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 2014, 5, 3477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, H.T.; Boucrot, E. Membrane curvature at a glance. J. Cell Sci. 2015, 128, 1065–1070. [Google Scholar] [CrossRef] [Green Version]
- Charrin, S.; Jouannet, S.; Boucheix, C.; Rubinstein, E. Tetraspanins at a glance. J. Cell Sci. 2014, 127, 3641–3648. [Google Scholar] [CrossRef] [Green Version]
- Charrin, S.; Manié, S.; Thiele, C.; Billard, M.; Gerlier, D.; Boucheix, C.; Rubinstein, E. A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol. 2003, 33, 2479–2489. [Google Scholar] [CrossRef]
- Termini, C.M.; Gillette, J.M. Tetraspanins Function as Regulators of Cellular Signaling. Front. Cell Dev. Biol. 2017, 5, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmerman, B.; Kelly, B.; McMillan, B.J.; Seegar, T.C.M.; Dror, R.O.; Kruse, A.C.; Blacklow, S.C. Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket. Cell 2016, 167, 1041–1051. [Google Scholar] [CrossRef] [Green Version]
- Chivet, M.; Javalet, C.; Laulagnier, K.; Blot, B.; Hemming, F.J.; Sadoul, R. Exosomes secreted by cortical neurons upon glutamatergic synapse activation specifically interact with neurons. J. Extracell. Vesicles 2014, 3, 24722. [Google Scholar] [CrossRef] [Green Version]
- Laulagnier, K.; Javalet, C.; Hemming, F.J.; Sadoul, R. Purification and Analysis of Exosomes Released by Mature Cortical Neurons Following Synaptic Activation. Methods Mol. Biol. 2017, 1545, 129–138. [Google Scholar] [CrossRef]
- Song, Y.; Li, Z.; He, T.; Qu, M.; Jiang, L.; Li, W.; Shi, X.; Pan, J.; Zhang, L.; Wang, Y.; et al. M2 microglia-derived exosomes protect the mouse brain from ischemia-reperfusion injury via exosomal miR-124. Theranostics 2019, 9, 2910–2923. [Google Scholar] [CrossRef]
- Li, D.; Huang, S.; Yin, Z.; Zhu, J.; Ge, X.; Han, Z.; Tan, J.; Zhang, S.; Zhao, J.; Chen, F.; et al. Increases in miR-124-3p in Microglial Exosomes Confer Neuroprotective Effects by Targeting FIP200-Mediated Neuronal Autophagy Following Traumatic Brain Injury. Neurochem. Res. 2019, 44, 1903–1923. [Google Scholar] [CrossRef]
- Gao, G.; Zhao, S.; Xia, X.; Li, C.; Li, C.; Ji, C.; Sheng, S.; Tang, Y.; Zhu, J.; Wang, Y.; et al. Glutaminase C Regulates Microglial Activation and Pro-inflammatory Exosome Release: Relevance to the Pathogenesis of Alzheimer’s Disease. Front. Cell. Neurosci. 2019, 13, 264. [Google Scholar] [CrossRef] [PubMed]
- Gabrielli, M.; Battista, N.; Riganti, L.; Prada, I.; Antonucci, F.; Cantone, L.; Matteoli, M.; Maccarrone, M.; Verderio, C. Active endocannabinoids are secreted on extracellular membrane vesicles. EMBO Rep. 2015, 16, 213–220. [Google Scholar] [CrossRef] [Green Version]
- Massenzio, F.; Peña-Altamira, E.; Petralla, S.; Virgili, M.; Zuccheri, G.; Miti, A.; Polazzi, E.; Mengoni, I.; Piffaretti, D.; Monti, B. Microglial overexpression of fALS-linked mutant SOD1 induces SOD1 processing impairment, activation and neurotoxicity and is counteracted by the autophagy inducer trehalose. Biochim. Biophys. Acta 2018, 2864, 3771–3785. [Google Scholar] [CrossRef]
- Glebov, K.; Löchner, M.; Jabs, R.; Lau, T.; Merkel, O.; Schloss, P.; Steinhäuser, C.; Walter, J. Serotonin stimulates secretion of exosomes from microglia cells. Glia 2015, 63, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Qian, B.; Kong, X.; Hao, J.; Ye, Y.; Yang, K.; Xu, T.; Zhang, F. A20 protects neuronal apoptosis stimulated by lipopolysaccharide-induced microglial exosomes. Neurosci. Lett. 2019, 712, 134480. [Google Scholar] [CrossRef]
- Ge, X.; Guo, M.; Hu, T.; Li, W.; Huang, S.; Yin, Z.; Li, Y.; Chen, F.; Zhu, L.; Kang, C.; et al. Increased Microglial Exosomal miR-124-3p Alleviates Neurodegeneration and Improves Cognitive Outcome after rmTBI. Mol. Ther. 2020, 28, 503–522. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Wu, Y.; Hu, Z.; Sun, L.; Dou, G.; Zhang, Z.; Wang, H.; Guo, C.; Wang, Y. Exosomes from Microglia Attenuate Photoreceptor Injury and Neovascularization in an Animal Model of Retinopathy of Prematurity. Mol. Ther.-Nucleic Acids 2019, 16, 778–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; Ge, X.; Yu, J.; Han, Z.; Yin, Z.; Li, Y.; Chen, F.; Wang, H.; Zhang, J.; Lei, P. Increased miR-124-3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB J. 2018, 32, 512–528. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Chen, C.; Huang, J.; Tian, Y.; Xie, X.; Yang, P.; Wu, M.; Tang, C.; Zhang, W. Nicotinamide phosphoribosyltransferase secreted from microglia via exosome during ischemic injury. J. Neurochem. 2019, 150, 723–737. [Google Scholar] [CrossRef]
- Gao, G.; Li, C.; Zhu, J.; Wang, Y.; Huang, Y.; Zhao, S.; Sheng, S.; Song, Y.; Ji, C.; Li, C.; et al. Glutaminase 1 Regulates Neuroinflammation After Cerebral Ischemia Through Enhancing Microglial Activation and Pro-Inflammatory Exosome Release. Front. Immunol. 2020, 11, 161. [Google Scholar] [CrossRef] [Green Version]
- Bakhti, M.; Winter, C. Inhibition of Myelin Membrane Sheath Formation by Oligodendrocyte-derived Exosome-like Vesicles. J. Biol. Chem. 2011, 286, 787–796. [Google Scholar] [CrossRef] [Green Version]
- Fitzner, D.; Schnaars, M.; van Rossum, D.; Krishnamoorthy, G.; Dibaj, P.; Bakhti, M.; Regen, T.; Hanisch, U.-K.; Simons, M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J. Cell Sci. 2011, 124, 447–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Shi, M.; Stewart, T.; Fernagut, P.-O.; Huang, Y.; Tian, C.; Dehay, B.; Atik, A.; Yang, D.; De Giorgi, F.; et al. Reduced oligodendrocyte exosome secretion in multiple system atrophy involves SNARE dysfunction. Brain 2020, 143, 1780–1797. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Verrilli, M.A.; Picou, F.; Court, F.A. Schwann cell-derived exosomes enhance axonal regeneration in the peripheral nervous system. Glia 2013, 61, 1795–1806. [Google Scholar] [CrossRef] [PubMed]
- Ching, R.C.; Wiberg, M.; Kingham, P.J. Schwann cell-like differentiated adipose stem cells promote neurite outgrowth via secreted exosomes and RNA transfer. Stem Cell Res. Ther. 2018, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Hu, M.; He, S.; Li, B.; Liu, C.; Min, J.; Hong, L. Effects of RSC96 Schwann Cell-Derived Exosomes on Proliferation, Senescence, and Apoptosis of Dorsal Root Ganglion Cells In Vitro. Med. Sci. Monit. 2018, 24, 7841–7849. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Fan, B.; Ding, H.; Liu, Y.; Tang, H.; Pan, D.; Shi, J.; Zheng, P.; Shi, H.; Wu, H.; et al. Proteomics analysis of Schwann cell-derived exosomes: A novel therapeutic strategy for central nervous system injury. Mol. Cell. Biochem. 2019, 457, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Chopp, M.; Szalad, A.; Lu, X.; Zhang, Y.; Wang, X.; Cepparulo, P.; Lu, M.; Li, C.; Zhang, Z.G. Exosomes Derived From Schwann Cells Ameliorate Peripheral Neuropathy in Type 2 Diabetic Mice. Diabetes 2020, 69, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Chopp, M.; Wang, L.; Lu, X.; Szalad, A.; Zhang, Z.G. Exosomes derived from high-glucose-stimulated Schwann cells promote development of diabetic peripheral neuropathy. FASEB J. 2018, 32, 6911–6922. [Google Scholar] [CrossRef] [Green Version]
- Abdullah, M.; Takase, H.; Nunome, M.; Enomoto, H.; Ito, J.; Gong, J.-S.; Michikawa, M. Amyloid-β Reduces Exosome Release from Astrocytes by Enhancing JNK Phosphorylation. J. Alzheimer’s Dis. 2016, 53, 1433–1441. [Google Scholar] [CrossRef]
- Hong, Y.; Zhao, T.; Li, X.-J.; Li, S. Mutant Huntingtin Inhibits αB-Crystallin Expression and Impairs Exosome Secretion from Astrocytes. J. Neurosci. 2017, 37, 9550–9563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jovičić, A.; Gitler, A.D. Distinct repertoires of microRNAs present in mouse astrocytes compared to astrocyte-secreted exosomes. PLoS ONE 2017, 12, e0171418. [Google Scholar] [CrossRef] [PubMed]
- Pužar Dominkuš, P.; Ferdin, J.; Plemenitaš, A.; Peterlin, B.M.; Lenassi, M. Nef is secreted in exosomes from Nef.GFP-expressing and HIV-1-infected human astrocytes. J. Neurovirol. 2017, 23, 713–724. [Google Scholar] [CrossRef]
- Ma, C.; Chen, H.; Zhang, S.; Yan, Y.; Wu, R.; Wang, Y.; Liu, Y.; Yang, L.; Liu, M. Exosomal and extracellular HMGB1 have opposite effects on SASH1 expression in rat astrocytes and glioma C6 cells. Biochem. Biophys. Res. Commun. 2019, 518, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Venturini, A.; Passalacqua, M.; Pelassa, S.; Pastorino, F.; Tedesco, M.; Cortese, K.; Gagliani, M.C.; Leo, G.; Maura, G.; Guidolin, D.; et al. Exosomes from astrocyte processes: Signaling to neurons. Front. Pharmacol. 2019, 10, 1452. [Google Scholar] [CrossRef] [Green Version]
- Xin, H.; Wang, F.; Li, Y.; Lu, Q.-E.; Cheung, W.L.; Zhang, Y.; Zhang, Z.G.; Chopp, M. Secondary Release of Exosomes from Astrocytes Contributes to the Increase in Neural Plasticity and Improvement of Functional Recovery after Stroke in Rats Treated with Exosomes Harvested from MicroRNA 133b-Overexpressing Multipotent Mesenchymal Stromal Ce. Cell Transplant. 2017, 26, 243–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lachenal, G.; Pernet-Gallay, K.; Chivet, M.; Hemming, F.J.; Belly, A.; Bodon, G.; Blot, B.; Haase, G.; Goldberg, Y.; Sadoul, R. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol. Cell. Neurosci. 2011, 46, 409–418. [Google Scholar] [CrossRef] [Green Version]
- Pinto, S.; Cunha, C.; Barbosa, M.; Vaz, A.R.; Brites, D. Exosomes from NSC-34 cells transfected with hSOD1-G93A are enriched in mir-124 and drive alterations in microglia phenotype. Front. Neurosci. 2017, 11, 273. [Google Scholar] [CrossRef] [Green Version]
- Le Gall, L.; Ouandaogo, Z.G.; Anakor, E.; Connolly, O.; Butler Browne, G.; Laine, J.; Duddy, W.; Duguez, S. Optimized method for extraction of exosomes from human primary muscle cells. Skelet. Muscle 2020, 10, 20. [Google Scholar] [CrossRef]
- Le Gall, L.; Duddy, W.J.; Martinat, C.; Mariot, V.; Connolly, O.; Milla, V.; Anakor, E.; Ouandaogo, Z.G.; Millecamps, S.; Lainé, J.; et al. Muscle cells of sporadic ALS patients secrete neurotoxic vesicles. medRxiv 2021. [Google Scholar] [CrossRef]
- Forterre, A.; Jalabert, A.; Berger, E.; Baudet, M.; Chikh, K.; Errazuriz, E.; De Larichaudy, J.; Chanon, S.; Weiss-Gayet, M.; Hesse, A.-M.; et al. Proteomic analysis of C2C12 myoblast and myotube exosome-like vesicles: A new paradigm for myoblast-myotube cross talk? PLoS ONE 2014, 9, e84153. [Google Scholar] [CrossRef]
- Le Bihan, M.-C.; Bigot, A.; Jensen, S.S.; Dennis, J.; Rogowska-Wrzesinska, A.; Lainé, J.; Gache, V.; Furling, D.; Jensen, O.N.; Voit, T.; et al. In-depth analysis of the secretome identifies three major independent secretory pathways in differentiating human myoblasts. J. Proteom. 2012, 77, 344–356. [Google Scholar] [CrossRef]
- Choi, J.S.; Yoon, H.I.; Lee, K.S.; Choi, Y.C.; Yang, S.H.; Kim, I.S.; Cho, Y.W. Exosomes from differentiating human skeletal muscle cells trigger myogenesis of stem cells and provide biochemical cues for skeletal muscle regeneration. J. Control. Release 2016, 222, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Jalabert, A.; Vial, G.; Guay, C.; Wiklander, O.P.B.; Nordin, J.Z.; Aswad, H.; Forterre, A.; Meugnier, E.; Pesenti, S.; Regazzi, R.; et al. Exosome-like vesicles released from lipid-induced insulin-resistant muscles modulate gene expression and proliferation of beta recipient cells in mice. Diabetologia 2016, 59, 1049–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aswad, H.; Forterre, A.; Wiklander, O.P.B.; Vial, G.; Danty-Berger, E.; Jalabert, A.; Lamazière, A.; Meugnier, E.; Pesenti, S.; Ott, C.; et al. Exosomes participate in the alteration of muscle homeostasis during lipid-induced insulin resistance in mice. Diabetologia 2014, 57, 2155–2164. [Google Scholar] [CrossRef]
- Nie, Y.; Sato, Y.; Garner, R.T.; Kargl, C.; Wang, C.; Kuang, S.; Gilpin, C.J.; Gavin, T.P. Skeletal muscle-derived exosomes regulate endothelial cell functions via reactive oxygen species-activated nuclear factor-κB signalling. Exp. Physiol. 2019, 104, 1262–1273. [Google Scholar] [CrossRef]
- Kim, S.; Lee, M.-J.; Choi, J.-Y.; Park, D.-H.; Kwak, H.-B.; Moon, S.; Koh, J.-W.; Shin, H.-K.; Ryu, J.-K.; Park, C.-S.; et al. Roles of Exosome-Like Vesicles Released from Inflammatory C2C12 Myotubes: Regulation of Myocyte Differentiation and Myokine Expression. Cell. Physiol. Biochem. 2018, 48, 1829–1842. [Google Scholar] [CrossRef]
- Romancino, D.P.; Buffa, V.; Caruso, S.; Ferrara, I.; Raccosta, S.; Notaro, A.; Campos, Y.; Noto, R.; Martorana, V.; Cupane, A.; et al. Palmitoylation is a post-translational modification of Alix regulating the membrane organization of exosome-like small extracellular vesicles. Biochim. Biophys. Acta-Gen. Subj. 2018, 1862, 2879–2887. [Google Scholar] [CrossRef] [Green Version]
- Forterre, A.; Jalabert, A.; Chikh, K.; Pesenti, S.; Euthine, V.; Granjon, A.; Errazuriz, E.; Lefai, E.; Vidal, H.; Rome, S. Myotube-derived exosomal miRNAs downregulate Sirtuin1 in myoblasts during muscle cell differentiation. Cell Cycle 2014, 13, 78–89. [Google Scholar] [CrossRef] [Green Version]
- Bonifacino, J.S.; Glick, B.S. The Mechanisms of Vesicle Budding and Fusion. Cell 2004, 116, 153–166. [Google Scholar] [CrossRef] [Green Version]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, C.; Morohashi, Y.; Yoshimura, S.-I.; Manrique-Hoyos, N.; Jung, S.; Lauterbach, M.A.; Bakhti, M.; Grønborg, M.; Möbius, W.; Rhee, J.; et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J. Cell Biol. 2010, 189, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Zabeo, D.; Cvjetkovic, A.; Lässer, C.; Schorb, M.; Lötvall, J.; Höög, J.L. Exosomes purified from a single cell type have diverse morphology. J. Extracell. Vesicles 2017, 6, 1329476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poliakov, A.; Spilman, M.; Dokland, T.; Amling, C.L.; Mobley, J.A. Structural heterogeneity and protein composition of exosome-like vesicles (prostasomes) in human semen. Prostate 2009, 69, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Aalberts, M.; van Dissel-Emiliani, F.M.; van Adrichem, N.P.; van Wijnen, M.; Wauben, M.H.; Stout, T.A.; Stoorvogel, W. Identification of Distinct Populations of Prostasomes That Differentially Express Prostate Stem Cell Antigen, Annexin A1, and GLIPR2 in Humans1. Biol. Reprod. 2012, 86. [Google Scholar] [CrossRef]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.I.E.; Lehtiö, J.; EL Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef]
- Bobrie, A.; Colombo, M.; Krumeich, S.; Raposo, G.; Théry, C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles 2012, 1, 18397. [Google Scholar] [CrossRef]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Stolz, D.B.; Sullivan, M.L.G.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z.; et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [Google Scholar] [CrossRef] [Green Version]
- Smith, Z.J.; Lee, C.; Rojalin, T.; Carney, R.P.; Hazari, S.; Knudson, A.; Lam, K.; Saari, H.; Ibañez, E.L.; Viitala, T.; et al. Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J. Extracell. Vesicles 2015, 4, 28533. [Google Scholar] [CrossRef] [Green Version]
- Van Niel, G.; Raposo, G.; Candalh, C.; Boussac, M.; Hershberg, R.; Cerf-Bensussan, N.; Heyman, M. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology 2001, 121, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Wubbolts, R.; Leckie, R.S.; Veenhuizen, P.T.M.; Schwarzmann, G.; Möbius, W.; Hoernschemeyer, J.; Slot, J.-W.; Geuze, H.J.; Stoorvogel, W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003, 278, 10963–10972. [Google Scholar] [CrossRef] [Green Version]
- Miyanishi, M.; Tada, K.; Koike, M.; Uchiyama, Y.; Kitamura, T.; Nagata, S. Identification of Tim4 as a phosphatidylserine receptor. Nature 2007, 450, 435–439. [Google Scholar] [CrossRef] [Green Version]
- Keller, S.; Sanderson, M.P.; Stoeck, A.; Altevogt, P. Exosomes: From biogenesis and secretion to biological function. Immunol. Lett. 2006, 107, 102–108. [Google Scholar] [CrossRef] [PubMed]
- de Gassart, A.; Geminard, C.; Fevrier, B.; Raposo, G.; Vidal, M. Lipid raft-associated protein sorting in exosomes. Blood 2003, 102, 4336–4344. [Google Scholar] [CrossRef] [Green Version]
- Anand, S.; Samuel, M.; Kumar, S.; Mathivanan, S. Ticket to a bubble ride: Cargo sorting into exosomes and extracellular vesicles. Biochim. Biophys. Acta-Proteins Proteom. 2019, 12, 140203. [Google Scholar] [CrossRef]
- Van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A common pathway for a specialized function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef]
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Record, M.; Silvente-Poirot, S.; Poirot, M.; Wakelam, M.J.O. Extracellular vesicles: Lipids as key components of their biogenesis and functions. J. Lipid Res. 2018, 59, 1316–1324. [Google Scholar] [CrossRef] [Green Version]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [Green Version]
- Danzer, K.M.; Kranich, L.R.; Ruf, W.P.; Cagsal-Getkin, O.; Winslow, A.R.; Zhu, L.; Vanderburg, C.R.; McLean, P.J. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol. Neurodegener. 2012, 7, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villarroya-Beltri, C.; Baixauli, F.; Mittelbrunn, M.; Fernández-Delgado, I.; Torralba, D.; Moreno-Gonzalo, O.; Baldanta, S.; Enrich, C.; Guerra, S.; Sánchez-Madrid, F. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat. Commun. 2016, 7, 13588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, H.; Chen, Q.; Lin, L.; Sha, C.; Li, T.; Liu, Y.; Yin, X.; Xu, Y.; Chen, L.; Gao, W.; et al. Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 2020, 17, 163–177. [Google Scholar] [CrossRef]
- McAndrews, K.M.; Kalluri, R. Mechanisms associated with biogenesis of exosomes in cancer. Mol. Cancer 2019, 18, 62. [Google Scholar] [CrossRef]
- Turchinovich, A.; Drapkina, O.; Tonevitsky, A. Transcriptome of extracellular vesicles: State-of-the-art. Front. Immunol. 2019, 10, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinforma. 2015, 13, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Guescini, M.; Guidolin, D.; Vallorani, L.; Casadei, L.; Gioacchini, A.M.; Tibollo, P.; Battistelli, M.; Falcieri, E.; Battistin, L.; Agnati, L.F.; et al. C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp. Cell Res. 2010, 316, 1977–1984. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janas, T.; Janas, M.M.; Sapoń, K.; Janas, T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015, 589, 1391–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic micrornas regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef] [Green Version]
- Santangelo, L.; Giurato, G.; Cicchini, C.; Montaldo, C.; Mancone, C.; Tarallo, R.; Battistelli, C.; Alonzi, T.; Weisz, A.; Tripodi, M. The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Rep. 2016, 17, 799–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, J.X.; Lv, L.H.; Wan, Y.L.; Cao, Y.; Li, G.L.; Lin, H.M.; Zhou, R.; Shang, C.Z.; Cao, J.; He, H.; et al. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology 2015, 61, 1284–1294. [Google Scholar] [CrossRef]
- Jackson, C.E.; Scruggs, B.S.; Schaffer, J.E.; Hanson, P.I. Effects of Inhibiting VPS4 Support a General Role for ESCRTs in Extracellular Vesicle Biogenesis. Biophys. J. 2017, 113, 1342–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janas, T.; Janas, T.; Yarus, M. Specific RNA binding to ordered phospholipid bilayers. Nucleic Acids Res. 2006, 34, 2128–2136. [Google Scholar] [CrossRef] [Green Version]
- Picca, A.; Guerra, F.; Calvani, R.; Coelho-Junior, H.J.; Bossola, M.; Landi, F.; Bernabei, R.; Bucci, C.; Marzetti, E. Generation and Release of Mitochondrial-Derived Vesicles in Health, Aging and Disease. J. Clin. Med. 2020, 9, 1440. [Google Scholar] [CrossRef]
- Malkin, E.Z.; Bratman, S.V. Bioactive DNA from extracellular vesicles and particles. Cell Death Dis. 2020, 11, 584. [Google Scholar] [CrossRef]
- Lázaro-Ibáñez, E.; Lässer, C.; Shelke, G.V.; Crescitelli, R.; Jang, S.C.; Cvjetkovic, A.; García-Rodríguez, A.; Lötvall, J. DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J. Extracell. Vesicles 2019, 8, 1656993. [Google Scholar] [CrossRef] [Green Version]
- Németh, A.; Orgovan, N.; Sódar, B.W.; Osteikoetxea, X.; Pálóczi, K.; Szabó-Taylor, K.; Vukman, K.V.; Kittel, Á.; Turiák, L.; Wiener, Z.; et al. Antibiotic-induced release of small extracellular vesicles (exosomes) with surface-associated DNA. Sci. Rep. 2017, 7, 8202. [Google Scholar] [CrossRef]
- Février, B.; Raposo, G. Exosomes: Endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004, 16, 415–421. [Google Scholar] [CrossRef]
- Gross, J.C.; Chaudhary, V.; Bartscherer, K.; Boutros, M. Active Wnt proteins are secreted on exosomes. Nat. Cell Biol. 2012, 14, 1036–1045. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, A.; Bandari, S.K.; Liu, J.; Mobley, J.A.; Brown, E.E.; Sanderson, R.D. Fibronectin on the Surface of Myeloma Cell-derived Exosomes Mediates Exosome-Cell Interactions. J. Biol. Chem. 2016, 291, 1652–1663. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Ding, Q.; Yaqoob, U.; de Assuncao, T.M.; Verma, V.K.; Hirsova, P.; Cao, S.; Mukhopadhyay, D.; Huebert, R.C.; Shah, V.H. Exosome Adherence and Internalization by Hepatic Stellate Cells Triggers Sphingosine 1-Phosphate-dependent Migration. J. Biol. Chem. 2015, 290, 30684–30696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segura, E.; Guérin, C.; Hogg, N.; Amigorena, S.; Théry, C. CD8+ dendritic cells use LFA-1 to capture MHC-peptide complexes from exosomes in vivo. J. Immunol. 2007, 179, 1489–1496. [Google Scholar] [CrossRef] [Green Version]
- Nolte-’t Hoen, E.N.M.; Buschow, S.I.; Anderton, S.M.; Stoorvogel, W.; Wauben, M.H.M. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood J. Am. Soc. Hematol. 2009, 113, 1977–1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segura, E.; Nicco, C.; Lombard, B.; Véron, P.; Raposo, G.; Batteux, F.; Amigorena, S.; Théry, C. ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood 2005, 106, 216–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Näslund, T.I.; Paquin-Proulx, D.; Paredes, P.T.; Vallhov, H.; Sandberg, J.K.; Gabrielsson, S. Exosomes from breast milk inhibit HIV-1 infection of dendritic cells and subsequent viral transfer to CD4+ T cells. AIDS 2014, 28, 171–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clayton, A.; Turkes, A.; Dewitt, S.; Steadman, R.; Mason, M.D.; Hallett, M.B. Adhesion and signaling by B cell-derived exosomes: The role of integrins. FASEB J. 2004, 18, 977–979. [Google Scholar] [CrossRef] [PubMed]
- Rieu, S.; Géminard, C.; Rabesandratana, H.; Sainte-Marie, J.; Vidal, M. Exosomes released during reticulocyte maturation bind to fibronectin via integrin α4β1. Eur. J. Biochem. 2000, 267, 583–590. [Google Scholar] [CrossRef]
- Hanayama, R.; Tanaka, M.; Miwa, K.; Shinohara, A.; Iwamatsu, A.; Nagata, S. Identification of a factor that links apoptotic cells to phagocytes. Nature 2002, 417, 182–187. [Google Scholar] [CrossRef]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef]
- Hao, S.; Bai, O.; Li, F.; Yuan, J.; Laferte, S.; Xiang, J. Mature dendritic cells pulsed with exosomes stimulate efficient cytotoxic T-lymphocyte responses and antitumour immunity. Immunology 2007, 120, 90–102. [Google Scholar] [CrossRef]
- Barrès, C.; Blanc, L.; Bette-Bobillo, P.; André, S.; Mamoun, R.; Gabius, H.-J.; Vidal, M. Galectin-5 is bound onto the surface of rat reticulocyte exosomes and modulates vesicle uptake by macrophages. Blood 2010, 115, 696–705. [Google Scholar] [CrossRef] [Green Version]
- Klibi, J.; Niki, T.; Riedel, A.; Pioche-Durieu, C.; Souquere, S.; Rubinstein, E.; Moulec, S.L.; Guigay, J.; Hirashima, M.; Guemira, F.; et al. Blood diffusion and Th1-suppressive effects of galectin-9–containing exosomes released by Epstein-Barr virus–infected nasopharyngeal carcinoma cells. Blood 2009, 113, 1957–1966. [Google Scholar] [CrossRef] [Green Version]
- Rana, S.; Claas, C.; Kretz, C.C.; Nazarenko, I.; Zoeller, M. Activation-induced internalization differs for the tetraspanins CD9 and Tspan8: Impact on tumor cell motility. Int. J. Biochem. Cell Biol. 2011, 43, 106–119. [Google Scholar] [CrossRef]
- Koumangoye, R.B.; Sakwe, A.M.; Goodwin, J.S.; Patel, T.; Ochieng, J. Detachment of breast tumor cells induces rapid secretion of exosomes which subsequently mediate cellular adhesion and spreading. PLoS ONE 2011, 6, e24234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saunderson, S.C.; Dunn, A.C.; Crocker, P.R.; McLellan, A.D. CD169 mediates the capture of exosomes in spleen and lymph node. Blood 2014, 123, 208–216. [Google Scholar] [CrossRef]
- Hung, W.-T.; Navakanitworakul, R.; Khan, T.; Zhang, P.; Davis, J.S.; McGinnis, L.K.; Christenson, L.K. Stage-specific follicular extracellular vesicle uptake and regulation of bovine granulosa cell proliferation. Biol. Reprod. 2017, 97, 644–655. [Google Scholar] [CrossRef]
- Hazan-Halevy, I.; Rosenblum, D.; Weinstein, S.; Bairey, O.; Raanani, P.; Peer, D. Cell-specific uptake of mantle cell lymphoma-derived exosomes by malignant and non-malignant B-lymphocytes. Cancer Lett. 2015, 364, 59–69. [Google Scholar] [CrossRef] [Green Version]
- Escrevente, C.; Keller, S.; Altevogt, P.; Costa, J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011, 11, 108. [Google Scholar] [CrossRef] [Green Version]
- Bretz, N.P.; Ridinger, J.; Rupp, A.-K.; Rimbach, K.; Keller, S.; Rupp, C.; Marmé, F.; Umansky, L.; Umansky, V.; Eigenbrod, T.; et al. Body fluid exosomes promote secretion of inflammatory cytokines in monocytic cells via Toll-like receptor signaling. J. Biol. Chem. 2013, 288, 36691–36702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [Green Version]
- Hakulinen, J.; Junnikkala, S.; Sorsa, T.; Meri, S. Complement inhibitor membrane cofactor protein (MCP; CD46) is constitutively shed from cancer cell membranes in vesicles and converted by a metalloproteinase to a functionally active soluble form. Eur. J. Immunol. 2004, 34, 2620–2629. [Google Scholar] [CrossRef]
- Prada, I.; Meldolesi, J. Binding and fusion of extracellular vesicles to the plasma membrane of their cell targets. Int. J. Mol. Sci. 2016, 17, 1296. [Google Scholar] [CrossRef] [Green Version]
- Tian, T.; Zhu, Y.-L.; Zhou, Y.-Y.; Liang, G.-F.; Wang, Y.-Y.; Hu, F.-H.; Xiao, Z.-D. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanbo, A.; Kawanishi, E.; Yoshida, R.; Yoshiyama, H. Exosomes Derived from Epstein-Barr Virus-Infected Cells Are Internalized via Caveola-Dependent Endocytosis and Promote Phenotypic Modulation in Target Cells. J. Virol. 2013, 87, 10334–10347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polanco, J.C.; Li, C.; Durisic, N.; Sullivan, R.; Götz, J. Exosomes taken up by neurons hijack the endosomal pathway to spread to interconnected neurons. Acta Neuropathol. Commun. 2018, 6, 10. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Su, H.; Li, J.; Lyon, C.; Tang, W.; Wan, M.; Hu, T.Y. Clinical applications of exosome membrane proteins. Precis. Clin. Med. 2020, 3, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Oluwole, S.O.A.; Yao, Y.; Conradi, S.; Kristensson, K.; Karlsson, H. Elevated levels of transcripts encoding a human retroviral envelope protein (syncytin) in muscles from patients with motor neuron disease. Amyotroph. Lateral Scler. 2007, 8, 67–72. [Google Scholar] [CrossRef]
- Bhat, R.K.; Ellestad, K.K.; Wheatley, B.M.; Warren, R.; Holt, R.A.; Power, C. Age- and disease-dependent HERV-W envelope allelic variation in brain: Association with neuroimmune gene expression. PLoS ONE 2011, 6, e19176. [Google Scholar] [CrossRef] [PubMed]
- Ben-Zvi, A.; Lacoste, B.; Kur, E.; Andreone, B.J.; Mayshar, Y.; Yan, H.; Gu, C. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature 2014, 509, 507–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidoryk-Wegrzynowicz, M. Impairment of glutamine/glutamate-γ-aminobutyric acid cycle in manganese toxicity in the central nervous system. Folia Neuropathol. 2014, 52, 377–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, Z.; Kamboj, A.; Anderson, C.M. Functional and immunocytochemical characterization of D-serine transporters in cortical neuron and astrocyte cultures. J. Neurosci. Res. 2009, 87, 2520–2530. [Google Scholar] [CrossRef] [PubMed]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH Is a Key Factor for Exosome Traffic in Tumor Cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [Green Version]
- Theparambil, S.M.; Hosford, P.S.; Ruminot, I.; Kopach, O.; Reynolds, J.R.; Sandoval, P.Y.; Rusakov, D.A.; Barros, L.F.; Gourine, A.V. Astrocytes regulate brain extracellular pH via a neuronal activity-dependent bicarbonate shuttle. Nat. Commun. 2020, 11, 5073. [Google Scholar] [CrossRef]
- Morelli, A.E.; Larregina, A.T.; Shufesky, W.J.; Sullivan, M.L.G.; Stolz, D.B.; Papworth, G.D.; Zahorchak, A.F.; Logar, A.J.; Wang, Z.; Watkins, S.C.; et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 2004, 104, 3257–3266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirchhausen, T. Clathrin. Annu. Rev. Biochem. 2000, 69, 699–727. [Google Scholar] [CrossRef] [PubMed]
- Kaksonen, M.; Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef]
- Anderson, R.G.W. The caveolae membrane system. Annu. Rev. Biochem. 1998, 67, 199–225. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Toufaily, C.; Annabi, B. Caveolin and cavin family members: Dual roles in cancer. Biochimie 2014, 107 Pt B, 188–202. [Google Scholar] [CrossRef]
- Horibe, S.; Tanahashi, T.; Kawauchi, S.; Murakami, Y.; Rikitake, Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer 2018, 18, 47. [Google Scholar] [CrossRef] [Green Version]
- Kerr, M.C.; Teasdale, R.D. Defining Macropinocytosis. Traffic 2009, 10, 364–371. [Google Scholar] [CrossRef]
- Feng, D.; Zhao, W.-L.; Ye, Y.-Y.; Bai, X.-C.; Liu, R.-Q.; Chang, L.-F.; Zhou, Q.; Sui, S.-F. Cellular Internalization of Exosomes Occurs Through Phagocytosis. Traffic 2010, 11, 675–687. [Google Scholar] [CrossRef]
- Doherty, G.J.; McMahon, H.T. Mechanisms of Endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svensson, K.J.; Christianson, H.C.; Wittrup, A.; Bourseau-Guilmain, E.; Lindqvist, E.; Svensson, L.M.; Mörgelin, M.; Belting, M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J. Biol. Chem. 2013, 288, 17713–17724. [Google Scholar] [CrossRef] [Green Version]
- El-Sayed, A.; Harashima, H. Endocytosis of gene delivery vectors: From clathrin-dependent to lipid raft-mediated endocytosis. Mol. Ther. 2013, 21, 1118–1130. [Google Scholar] [CrossRef] [Green Version]
- McKelvey, K.J.; Powell, K.L.; Ashton, A.W.; Morris, J.M.; McCracken, S.A. Exosomes: Mechanisms of Uptake. J. Circ. Biomark. 2015, 4, 7. [Google Scholar] [CrossRef] [Green Version]
- Gruenberg, J.; van der Goot, F.G. Mechanisms of pathogen entry through the endosomal compartments. Nat. Rev. Mol. Cell Biol. 2006, 7, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Men, Y.; Yelick, J.; Jin, S.; Tian, Y.; Chiang, M.S.R.; Higashimori, H.; Brown, E.; Jarvis, R.; Yang, Y. Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat. Commun. 2019, 10, 4136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.Y.; Huang, C.M.; Hung, Y.F.; Hsueh, Y.P. The microRNAs Let7c and miR21 are recognized by neuronal Toll-like receptor 7 to restrict dendritic growth of neurons. Exp. Neurol. 2015, 269, 202–212. [Google Scholar] [CrossRef]
- Goldie, B.J.; Dun, M.D.; Lin, M.; Smith, N.D.; Verrills, N.M.; Dayas, C.V.; Cairns, M.J. Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 2014, 42, 9195–9208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; Shin, S.M.; Zhong, P.; Kim, H.T.; Kim, D.I.; Kim, J.M.; Do Heo, W.; Kim, D.W.; Yeo, C.Y.; Kim, C.H.; et al. Reciprocal control of excitatory synapse numbers by Wnt and Wnt inhibitor PRR7 secreted on exosomes. Nat. Commun. 2018, 9, 3434. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Koutras, C.; Donnelier, J.; Alshehri, M.; Fotouhi, M.; Girard, M.; Casha, S.; McPherson, P.S.; Robbins, S.M.; Braun, J.E.A. Neurons Export Extracellular Vesicles Enriched in Cysteine String Protein and Misfolded Protein Cargo. Sci. Rep. 2017, 7, 956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahrini, I.; Song, J.H.; Diez, D.; Hanayama, R. Neuronal exosomes facilitate synaptic pruning by up-regulating complement factors in microglia. Sci. Rep. 2015, 5, 7989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morel, L.; Regan, M.; Higashimori, H.; Ng, S.K.; Esau, C.; Vidensky, S.; Rothstein, J.; Yang, Y. Neuronal exosomal mirna-dependent translational regulation of astroglial glutamate transporter glt1. J. Biol. Chem. 2013, 288, 7105–7116. [Google Scholar] [CrossRef] [Green Version]
- Goncalves, M.B.; Malmqvist, T.; Clarke, E.; Hubens, C.J.; Grist, J.; Hobbs, C.; Trigo, D.; Risling, M.; Angeria, M.; Damberg, P.; et al. Neuronal RARβ signaling modulates PTEN activity directly in neurons and via exosome transfer in astrocytes to prevent glial scar formation and induce spinal cord regeneration. J. Neurosci. 2015, 35, 15731–15745. [Google Scholar] [CrossRef] [PubMed]
- Guitart, K.; Loers, G.; Buck, F.; Bork, U.; Schachner, M.; Kleene, R. Improvement of neuronal cell survival by astrocyte-derived exosomes under hypoxic and ischemic conditions depends on prion protein. Glia 2016, 64, 896–910. [Google Scholar] [CrossRef]
- Pascua-Maestro, R.; González, E.; Lillo, C.; Ganfornina, M.D.; Falcón-Pérez, J.M.; Sanchez, D. Extracellular vesicles secreted by astroglial cells transport apolipoprotein D to neurons and mediate neuronal survival upon oxidative stress. Front. Cell. Neurosci. 2019, 12, 526. [Google Scholar] [CrossRef]
- Chen, W.; Zheng, P.; Hong, T.; Wang, Y.; Liu, N.; He, B.; Zou, S.; Ren, D.; Duan, J.; Zhao, L.; et al. Astrocytes-derived exosomes induce neuronal recovery after traumatic brain injury via delivering gap junction alpha 1-20 k. J. Tissue Eng. Regen. Med. 2020, 14, 412–423. [Google Scholar] [CrossRef]
- Romancino, D.P.; Paterniti, G.; Campos, Y.; De Luca, A.; Di Felice, V.; D’Azzo, A.; Bongiovanni, A. Identification and characterization of the nano-sized vesicles released by muscle cells. FEBS Lett. 2013, 587, 1379–1384. [Google Scholar] [CrossRef] [Green Version]
- Madison, R.D.; McGee, C.; Rawson, R.; Robinson, G.A. Extracellular vesicles from a muscle cell line (C2C12) enhance cell survival and neurite outgrowth of a motor neuron cell line (NSC-34). J. Extracell. Vesicles 2014, 3, 22865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baci, D.; Chirivì, M.; Pace, V.; Maiullari, F.; Milan, M.; Rampin, A.; Somma, P.; Presutti, D.; Garavelli, S.; Bruno, A.; et al. Extracellular Vesicles from Skeletal Muscle Cells Efficiently Promote Myogenesis in Induced Pluripotent Stem Cells. Cells 2020, 9, 1527. [Google Scholar] [CrossRef] [PubMed]
- Thorley, M.; Malatras, A.; Duddy, W.J.; Le Gall, L.; Mouly, V.; Butler Browne, G.; Duguez, S.M.-R. Changes in communication between muscle stem cells and their environment with aging. J. Neuromuscul. Dis. 2015, 2, 205–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohashi, M.; Korsakova, E.; Allen, D.; Lee, P.; Fu, K.; Vargas, B.S.; Cinkornpumin, J.; Salas, C.; Park, J.C.; Germanguz, I.; et al. Loss of MECP2 Leads to Activation of P53 and Neuronal Senescence. Stem Cell Rep. 2018, 10, 1453–1463. [Google Scholar] [CrossRef] [PubMed]
- Bigot, A.; Duddy, W.J.; Ouandaogo, Z.G.; Negroni, E.; Mariot, V.; Ghimbovschi, S.; Harmon, B.; Wielgosik, A.; Loiseau, C.; Devaney, J.; et al. Age-Associated Methylation Suppresses SPRY1, Leading to a Failure of Re-quiescence and Loss of the Reserve Stem Cell Pool in Elderly Muscle. Cell Rep. 2015, 13, 1172–1182. [Google Scholar] [CrossRef] [Green Version]
- Barberi, L.; Scicchitano, B.M.; De Rossi, M.; Bigot, A.; Duguez, S.; Wielgosik, A.; Stewart, C.; McPhee, J.; Conte, M.; Narici, M.; et al. Age-dependent alteration in muscle regeneration: The critical role of tissue niche. Biogerontology 2013, 14, 273–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franceschi, C.; Capri, M.; Monti, D.; Giunta, S.; Olivieri, F.; Sevini, F.; Panourgia, M.P.; Invidia, L.; Celani, L.; Scurti, M.; et al. Inflammaging and anti-inflammaging: A systemic perspective on aging and longevity emerged from studies in humans. Mech. Ageing Dev. 2007, 128, 92–105. [Google Scholar] [CrossRef]
- Pandya, V.A.; Patani, R. Decoding the relationship between ageing and amyotrophic lateral sclerosis: A cellular perspective. Brain 2020, 143, 1057–1072. [Google Scholar] [CrossRef] [PubMed]
- Alibhai, F.J.; Lim, F.; Yeganeh, A.; DiStefano, P.V.; Binesh-Marvasti, T.; Belfiore, A.; Wlodarek, L.; Gustafson, D.; Millar, S.; Li, S.-H.; et al. Cellular senescence contributes to age-dependent changes in circulating extracellular vesicle cargo and function. Aging Cell 2020, 19, e13103. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.L.; Lukas, T.J.; Yuan, M.; Du, N.; Tso, M.O.; Neufeld, A.H. Autophagy and exosomes in the aged retinal pigment epithelium: Possible relevance to drusen formation and age-related macular degeneration. PLoS ONE 2009, 4, e4160. [Google Scholar] [CrossRef] [Green Version]
- Takasugi, M.; Okada, R.; Takahashi, A.; Virya Chen, D.; Watanabe, S.; Hara, E. Small extracellular vesicles secreted from senescent cells promote cancer cell proliferation through EphA2. Nat. Commun. 2017, 8, 15729. [Google Scholar] [CrossRef]
- Takasugi, M. Emerging roles of extracellular vesicles in cellular senescence and aging. Aging Cell 2018, 17, e12734. [Google Scholar] [CrossRef] [PubMed]
- Wells, S.I.; Aronow, B.J.; Wise, T.M.; Williams, S.S.; Couget, J.A.; Howley, P.M. Transcriptome signature of irreversible senescence in human papillomavirus-positive cervical cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 7093–7098. [Google Scholar] [CrossRef] [Green Version]
- Fujii, M.; Kawai, Y.; Endoh, M.; Hossain, M.N.; Nakabayashi, K.; Ayusawa, D. Expression of RAB27B is up-regulated in senescent human cells. Mech. Ageing Dev. 2006, 127, 639–642. [Google Scholar] [CrossRef]
- Shamseddine, A.A.; Clarke, C.J.; Carroll, B.; Airola, M.V.; Mohammed, S.; Rella, A.; Obeid, L.M.; Hannun, Y.A. P53-dependent upregulation of neutral sphingomyelinase-2: Role in doxorubicin-induced growth arrest. Cell Death Dis. 2015, 6, e1947. [Google Scholar] [CrossRef] [Green Version]
- Hitomi, K.; Okada, R.; Loo, T.M.; Miyata, K.; Nakamura, A.J.; Takahashi, A. DNA Damage Regulates Senescence-Associated Extracellular Vesicle Release via the Ceramide Pathway to Prevent Excessive Inflammatory Responses. Int. J. Mol. Sci. 2020, 21, 3720. [Google Scholar] [CrossRef]
- Kadota, T.; Fujita, Y.; Yoshioka, Y.; Araya, J.; Kuwano, K.; Ochiya, T. Emerging role of extracellular vesicles as a senescence-associated secretory phenotype: Insights into the pathophysiology of lung diseases. Mol. Asp. Med. 2018, 60, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Hettinger, Z.R.; Kargl, C.K.; Shannahan, J.H.; Kuang, S.; Gavin, T.P. Extracellular vesicles released from stress-induced prematurely senescent myoblasts impair endothelial function and proliferation. Exp. Physiol. 2021, 106, 2083–2095. [Google Scholar] [CrossRef]
- Fulzele, S.; Mendhe, B.; Khayrullin, A.; Johnson, M.; Kaiser, H.; Liu, Y.; Isales, C.M.; Hamrick, M.W. Muscle-derived miR-34a increases with age in circulating extracellular vesicles and induces senescence of bone marrow stem cells. Aging (Albany NY) 2019, 11, 1791–1803. [Google Scholar] [CrossRef]
- Yang, C.P.; Yang, W.S.; Wong, Y.H.; Wang, K.H.; Teng, Y.C.; Chang, M.H.; Liao, K.H.; Nian, F.S.; Chao, C.C.; Tsai, J.W.; et al. Muscle atrophy-related myotube-derived exosomal microRNA in neuronal dysfunction: Targeting both coding and long noncoding RNAs. Aging Cell 2020, 19. [Google Scholar] [CrossRef] [PubMed]
- Fafián-Labora, J.A.; O’Loghlen, A. NF-κB/IKK activation by small extracellular vesicles within the SASP. Aging Cell 2021, 20, e13426. [Google Scholar] [CrossRef]
- Tan, D.B.A.; Armitage, J.; Teo, T.-H.; Ong, N.E.; Shin, H.; Moodley, Y.P. Elevated levels of circulating exosome in COPD patients are associated with systemic inflammation. Respir. Med. 2017, 132, 261–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberro, A.; Sáenz-Cuesta, M.; Muñoz-Culla, M.; Mateo-Abad, M.; Gonzalez, E.; Carrasco-Garcia, E.; Araúzo-Bravo, M.J.; Matheu, A.; Vergara, I.; Otaegui, D. Inflammaging and Frailty Status Do Not Result in an Increased Extracellular Vesicle Concentration in Circulation. Int. J. Mol. Sci. 2016, 17, 1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eitan, E.; Green, J.; Bodogai, M.; Mode, N.A.; Bæk, R.; Jørgensen, M.M.; Freeman, D.W.; Witwer, K.W.; Zonderman, A.B.; Biragyn, A.; et al. Age-Related Changes in Plasma Extracellular Vesicle Characteristics and Internalization by Leukocytes. Sci. Rep. 2017, 7, 1342. [Google Scholar] [CrossRef] [Green Version]
- Weilner, S.; Keider, V.; Winter, M.; Harreither, E.; Salzer, B.; Weiss, F.; Schraml, E.; Messner, P.; Pietschmann, P.; Hildner, F.; et al. Vesicular Galectin-3 levels decrease with donor age and contribute to the reduced osteo-inductive potential of human plasma derived extracellular vesicles. Aging (Albany NY) 2016, 8, 16–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Kim, M.S.; Jia, B.; Yan, J.; Zuniga-Hertz, J.P.; Han, C.; Cai, D. Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature 2017, 548, 52–57. [Google Scholar] [CrossRef]
- Terlecki-Zaniewicz, L.; Lämmermann, I.; Latreille, J.; Bobbili, M.R.; Pils, V.; Schosserer, M.; Weinmüllner, R.; Dellago, H.; Skalicky, S.; Pum, D.; et al. Small extracellular vesicles and their miRNA cargo are anti-apoptotic members of the senescence-associated secretory phenotype. Aging (Albany NY) 2018, 10, 1103–1132. [Google Scholar] [CrossRef]
- Luo, Z.; Feng, X.; Wang, H.; Xu, W.; Zhao, Y.; Ma, W.; Jiang, S.; Liu, D.; Huang, J.; Songyang, Z. Mir-23a induces telomere dysfunction and cellular senescence by inhibiting TRF2 expression. Aging Cell 2015, 14, 391–399. [Google Scholar] [CrossRef]
- Prattichizzo, F.; Giuliani, A.; Sabbatinelli, J.; Mensà, E.; De Nigris, V.; La Sala, L.; de Candia, P.; Olivieri, F.; Ceriello, A. Extracellular vesicles circulating in young organisms promote healthy longevity. J. Extracell. Vesicles 2019, 8, 1656044. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, M.; Satoh, A.; Lin, J.B.; Mills, K.F.; Sasaki, Y.; Rensing, N.; Wong, M.; Apte, R.S.; Imai, S.-I. Extracellular Vesicle-Contained eNAMPT Delays Aging and Extends Lifespan in Mice. Cell Metab. 2019, 30, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Mahairaki, V.; Bai, H.; Ding, Z.; Li, J.; Witwer, K.W.; Cheng, L. Highly Purified Human Extracellular Vesicles Produced by Stem Cells Alleviate Aging Cellular Phenotypes of Senescent Human Cells. Stem Cells 2019, 37, 779–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, G.; Xia, Y.; Zhang, J.; Chen, Y.; Yuan, J.; Niu, X.; Zhao, B.; Li, Q.; Wang, Y.; Deng, Z. ESC-sEVs Rejuvenate Senescent Hippocampal NSCs by Activating Lysosomes to Improve Cognitive Dysfunction in Vascular Dementia. Adv. Sci. 2020, 7, 1903330. [Google Scholar] [CrossRef] [Green Version]
- Fafián-Labora, J.A.; Rodríguez-Navarro, J.A.; O’Loghlen, A. Small Extracellular Vesicles Have GST Activity and Ameliorate Senescence-Related Tissue Damage. Cell Metab. 2020, 32, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wei, J.; Da Fonseca Ferreira, A.; Wang, H.; Zhang, L.; Zhang, Q.; Bellio, M.A.; Chu, X.-M.; Khan, A.; Jayaweera, D.; et al. Rejuvenation of Senescent Endothelial Progenitor Cells by Extracellular Vesicles Derived From Mesenchymal Stromal Cells. JACC. Basic Transl. Sci. 2020, 5, 1127–1141. [Google Scholar] [CrossRef]
- Dorronsoro, A.; Santiago, F.E.; Grassi, D.; Zhang, T.; Lai, R.C.; McGowan, S.J.; Angelini, L.; Lavasani, M.; Corbo, L.; Lu, A.; et al. Mesenchymal stem cell-derived extracellular vesicles reduce senescence and extend health span in mouse models of aging. Aging Cell 2021, 20, e13337. [Google Scholar] [CrossRef]
- Chai, A.; Withers, J.; Koh, Y.H.; Parry, K.; Bao, H.; Zhang, B.; Budnik, V.; Pennetta, G. hVAPB, the causative gene of a heterogeneous group of motor neuron diseases in humans, is functionally interchangeable with its Drosophila homologue DVAP-33A at the neuromuscular junction. Hum. Mol. Genet. 2008, 17, 266–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connolly, O.; Le Gall, L.; McCluskey, G.; Donaghy, C.G.; Duddy, W.J.; Duguez, S. A Systematic Review of Genotype–Phenotype Correlation across Cohorts Having Causal Mutations of Different Genes in ALS. J. Pers. Med. 2020, 10, 58. [Google Scholar] [CrossRef]
- Turner, M.R.; Al-Chalabi, A.; Chio, A.; Hardiman, O.; Kiernan, M.C.; Rohrer, J.D.; Rowe, J.; Seeley, W.; Talbot, K. Genetic screening in sporadic ALS and FTD. J. Neurol. Neurosurg. Psychiatry 2017, 88, 1042–1044. [Google Scholar] [CrossRef] [Green Version]
- Le Gall, L.; Anakor, E.; Connolly, O.; Vijayakumar, U.G.; Duddy, W.J.; Duguez, S.; Le Gall, L.; Anakor, E.; Connolly, O.; Vijayakumar, U.G.; et al. Molecular and Cellular Mechanisms Affected in ALS. J. Pers. Med. 2020, 10, 101. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Ma, M.; Teng, J.; Teng, R.K.F.; Zhou, S.; Yin, J.; Fonkem, E.; Huang, J.H.; Wu, E.; Wang, X. Exposure to ALS-FTD-CSF generates TDP-43 aggregates in glioblastoma cells through exosomes and TNTs-like structure. Oncotarget 2015, 6, 24178–24191. [Google Scholar] [CrossRef] [Green Version]
- Feiler, M.S.; Strobel, B.; Freischmidt, A.; Helferich, A.M.; Kappel, J.; Brewer, B.M.; Li, D.; Thal, D.R.; Walther, P.; Ludolph, A.C.; et al. TDP-43 is intercellularly transmitted across axon terminals. J. Cell Biol. 2015, 211, 897–911. [Google Scholar] [CrossRef]
- Westergard, T.; Jensen, B.K.; Wen, X.; Cai, J.; Kropf, E.; Iacovitti, L.; Pasinelli, P.; Trotti, D. Cell-to-Cell Transmission of Dipeptide Repeat Proteins Linked to C9orf72 -ALS/FTD. Cell Rep. 2016, 17, 645–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAlary, L.; Plotkin, S.S.; Yerbury, J.J.; Cashman, N.R. Prion-Like Propagation of Protein Misfolding and Aggregation in Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2019, 12, 272. [Google Scholar] [CrossRef] [Green Version]
- Gomes, C.; Keller, S.; Altevogt, P.; Costa, J. Evidence for secretion of Cu,Zn superoxide dismutase via exosomes from a cell model of amyotrophic lateral sclerosis. Neurosci. Lett. 2007, 428, 43–46. [Google Scholar] [CrossRef]
- Basso, M.; Pozzi, S.; Tortarolo, M.; Fiordaliso, F.; Bisighini, C.; Pasetto, L.; Spaltro, G.; Lidonnici, D.; Gensano, F.; Battaglia, E.; et al. Mutant copper-zinc superoxide dismutase (SOD1) induces protein secretion pathway alterations and exosome release in astrocytes: Implications for disease spreading and motor neuron pathology in amyotrophic lateral sclerosis. J. Biol. Chem. 2013, 288, 15699–15711. [Google Scholar] [CrossRef] [Green Version]
- Vaz, A.R.; Pinto, S.; Ezequiel, C.; Cunha, C.; Carvalho, L.A.; Moreira, R.; Brites, D. Phenotypic effects of wild-type and mutant SOD1 expression in n9 murine microglia at steady state, inflammatory and immunomodulatory conditions. Front. Cell. Neurosci. 2019, 13, 109. [Google Scholar] [CrossRef] [Green Version]
- Grad, L.I.; Yerbury, J.J.; Turner, B.J.; Guest, W.C.; Pokrishevsky, E.; O’Neill, M.A.; Yanai, A.; Silverman, J.M.; Zeineddine, R.; Corcoran, L.; et al. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. USA 2014, 111, 3620–3625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, J.S.; Fuentealba, R.A.; Miller, S.E.; Jackson, E.; Piwnica-Worms, D.; Baloh, R.H.; Weihl, C.C. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J. Cell Biol. 2009, 187, 875–888. [Google Scholar] [CrossRef]
- Lagier-Tourenne, C.; Polymenidou, M.; Cleveland, D.W. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010, 19, R46–R64. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, I.R.A.; Rademakers, R.; Neumann, M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010, 9, 995–1007. [Google Scholar] [CrossRef]
- Nonaka, T.; Masuda-Suzukake, M.; Arai, T.; Hasegawa, Y.; Akatsu, H.; Obi, T.; Yoshida, M.; Murayama, S.; Mann, D.M.A.; Akiyama, H.; et al. Prion-like Properties of Pathological TDP-43 Aggregates from Diseased Brains. Cell Rep. 2013, 4, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Iguchi, Y.; Eid, L.; Parent, M.; Soucy, G.; Bareil, C.; Riku, Y.; Kawai, K.; Takagi, S.; Yoshida, M.; Katsuno, M.; et al. Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain 2016, 139, 3187–3201. [Google Scholar] [CrossRef] [Green Version]
- Kamelgarn, M.; Chen, J.; Kuang, L.; Arenas, A.; Zhai, J.; Zhu, H.; Gal, J. Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS. Biochim. Biophys. Acta-Mol. Basis Dis. 2016, 1862, 2004–2014. [Google Scholar] [CrossRef]
- Leko, M.B.; Župunski, V.; Kirincich, J.; Smilović, D.; Hortobágyi, T.; Hof, P.R.; Šimić, G. Molecular mechanisms of neurodegeneration related to C9orf72 hexanucleotide repeat expansion. Behav. Neurol. 2019, 2019, 2909168. [Google Scholar] [CrossRef] [Green Version]
- Desdín-Micó, G.; Mittelbrunn, M. Role of exosomes in the protection of cellular homeostasis. Cell Adhes. Migr. 2017, 11, 127–134. [Google Scholar] [CrossRef] [Green Version]
- Grad, L.I.; Cashman, N.R. Prion-like activity of Cu/Zn superoxide dismutase. Prion 2014, 8, 33–41. [Google Scholar] [CrossRef]
- Kovanda, A.; Leonardis, L.; Zidar, J.; Koritnik, B.; Dolenc-Groselj, L.; Ristic Kovacic, S.; Curk, T.; Rogelj, B. Differential expression of microRNAs and other small RNAs in muscle tissue of patients with ALS and healthy age-matched controls. Sci. Rep. 2018, 8, 5609. [Google Scholar] [CrossRef]
- Emde, A.; Eitan, C.; Liou, L.; Libby, R.T.; Rivkin, N.; Magen, I.; Reichenstein, I.; Oppenheim, H.; Eilam, R.; Silvestroni, A.; et al. Dysregulated mi RNA biogenesis downstream of cellular stress and ALS -causing mutations: A new mechanism for ALS. EMBO J. 2015, 34, 2633–2651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koval, E.D.; Shaner, C.; Zhang, P.; du Maine, X.; Fischer, K.; Tay, J.; Chau, B.N.; Wu, G.F.; Miller, T.M. Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Hum. Mol. Genet. 2013, 22, 4127–4135. [Google Scholar] [CrossRef] [Green Version]
- Campos-Melo, D.; Droppelmann, C.A.; He, Z.; Volkening, K.; Strong, M.J. Altered microRNA expression profile in amyotrophic lateral sclerosis: A role in the regulation of NFL mRNA levels. Mol. Brain 2013, 6, 26. [Google Scholar] [CrossRef] [Green Version]
- De Felice, B.; Guida, M.; Guida, M.; Coppola, C.; De Mieri, G.; Cotrufo, R. A miRNA signature in leukocytes from sporadic amyotrophic lateral sclerosis. Gene 2012, 508, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Guan, Y.; Chen, Y.; Zhang, C.; Yu, L.; Gao, H.; Du, H.; Liu, B.; Wang, X. miRNA-9 expression is upregulated in the spinal cord of G93A-SOD1 transgenic mice. Int. J. Clin. Exp. Pathol. 2013, 6, 1826–1838. [Google Scholar] [PubMed]
- Hoye, M.L.; Koval, E.D.; Wegener, A.J.; Hyman, T.S.; Yang, C.; O’Brien, D.R.; Miller, R.L.; Cole, T.; Schoch, K.M.; Shen, T.; et al. MicroRNA profiling reveals marker of motor neuron disease in ALS models. J. Neurosci. 2017, 37, 5574–5586. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; Van Eijndhoven, M.A.J.; Hopmans, E.S.; Lindenberg, J.L.; De Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [Green Version]
- Varcianna, A.; Myszczynska, M.A.; Castelli, L.M.; O’Neill, B.; Kim, Y.; Talbot, J.; Nyberg, S.; Nyamali, I.; Heath, P.R.; Stopford, M.J.; et al. Micro-RNAs secreted through astrocyte-derived extracellular vesicles cause neuronal network degeneration in C9orf72 ALS. EBioMedicine 2019, 40, 626–635. [Google Scholar] [CrossRef] [Green Version]
- Dardiotis, E.; Aloizou, A.M.; Siokas, V.; Patrinos, G.P.; Deretzi, G.; Mitsias, P.; Aschner, M.; Tsatsakis, A. The Role of MicroRNAs in Patients with Amyotrophic Lateral Sclerosis. J. Mol. Neurosci. 2018, 66, 617–628. [Google Scholar] [CrossRef]
- Onyeagucha, B.C.; Mercado-Pimentel, M.E.; Hutchison, J.; Flemington, E.K.; Nelson, M.A. S100P/RAGE signaling regulates microRNA-155 expression via AP-1 activation in colon cancer. Exp. Cell Res. 2013, 319, 2081–2090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birger, A.; Ottolenghi, M.; Perez, L.; Reubinoff, B.; Behar, O. ALS-related human cortical and motor neurons survival is differentially affected by Sema3A article. Cell Death Dis. 2018, 9, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korner, S.; Boselt, S.; Wichmann, K.; Thau-Habermann, N.; Zapf, A.; Knippenberg, S.; Dengler, R.; Petri, S. The axon guidance protein semaphorin 3A is increased in the motor cortex of patients with amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 2016, 75, 326–333. [Google Scholar] [CrossRef] [Green Version]
- Conlan, R.S.; Pisano, S.; Oliveira, M.I.; Ferrari, M.; Mendes Pinto, I. Exosomes as Reconfigurable Therapeutic Systems. Trends Mol. Med. 2017, 23, 636–650. [Google Scholar] [CrossRef]
- Lee, M.; Ban, J.J.; Kim, K.Y.; Jeon, G.S.; Im, W.; Sung, J.J.; Kim, M. Adipose-derived stem cell exosomes alleviate pathology of amyotrophic lateral sclerosis in vitro. Biochem. Biophys. Res. Commun. 2016, 479, 434–439. [Google Scholar] [CrossRef]
- Calabria, E.; Scambi, I.; Bonafede, R.; Schiaffino, L.; Peroni, D.; Potrich, V.; Capelli, C.; Schena, F.; Mariotti, R. Ascs-exosomes recover coupling efficiency and mitochondrial membrane potential in an in vitro model of als. Front. Neurosci. 2019, 13, 1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonafede, R.; Scambi, I.; Peroni, D.; Potrich, V.; Boschi, F.; Benati, D.; Bonetti, B.; Mariotti, R. Exosome derived from murine adipose-derived stromal cells: Neuroprotective effect on in vitro model of amyotrophic lateral sclerosis. Exp. Cell Res. 2016, 340, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Bonafede, R.; Brandi, J.; Manfredi, M.; Scambi, I.; Schiaffino, L.; Merigo, F.; Turano, E.; Bonetti, B.; Marengo, E.; Cecconi, D.; et al. The Anti-Apoptotic Effect of ASC-Exosomes in an In Vitro ALS Model and Their Proteomic Analysis. Cells 2019, 8, 1087. [Google Scholar] [CrossRef] [Green Version]
- Venugopal, C.; Rai, K.S.; Pinnelli, V.B.; Kutty, B.M.; Dhanushkodi, A. Neuroprotection by Human Dental Pulp Mesenchymal Stem Cells: From Billions to Nano. Curr. Gene Ther. 2018, 18, 307–323. [Google Scholar] [CrossRef]
- Ludwig, N.; Whiteside, T.L.; Reichert, T.E. Challenges in exosome isolation and analysis in health and disease. Int. J. Mol. Sci. 2019, 20, 4684. [Google Scholar] [CrossRef] [Green Version]
- Venugopal, C.; Shamir, C.; Senthilkumar, S.; Babu, J.V.; Sonu, P.K.; Nishtha, K.J.; Rai, K.S.; Shobha, K.; Dhanushkodi, A. Dosage and Passage Dependent Neuroprotective Effects of Exosomes Derived from Rat Bone Marrow Mesenchymal Stem Cells: An In Vitro Analysis. Curr. Gene Ther. 2018, 17, 379–390. [Google Scholar] [CrossRef]
- Bonafede, R.; Turano, E.; Scambi, I.; Busato, A.; Bontempi, P.; Virla, F.; Schiaffino, L.; Marzola, P.; Bonetti, B.; Mariotti, R. ASC-exosomes ameliorate the disease progression in SOD1(G93A) murine model underlining their potential therapeutic use in human ALS. Int. J. Mol. Sci. 2020, 21, 3651. [Google Scholar] [CrossRef]
- Kooijmans, S.A.A.; Schiffelers, R.M.; Zarovni, N.; Vago, R. Modulation of tissue tropism and biological activity of exosomes and other extracellular vesicles: New nanotools for cancer treatment. Pharmacol. Res. 2016, 111, 487–500. [Google Scholar] [CrossRef]
- Lin, Y.; Lu, Y.; Li, X. Biological characteristics of exosomes and genetically engineered exosomes for the targeted delivery of therapeutic agents. J. Drug Target. 2020, 28, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef] [Green Version]
- Hung, M.E.; Leonard, J.N. Stabilization of exosome-targeting peptides via engineered glycosylation. J. Biol. Chem. 2015, 290, 8166–8172. [Google Scholar] [CrossRef] [Green Version]
- Murphy, D.E.; de Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular vesicle-based therapeutics: Natural versus engineered targeting and trafficking. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Singer, O.; Marr, R.A.; Rockenstein, E.; Crews, L.; Coufal, N.G.; Gage, F.H.; Verma, I.M.; Masliah, E. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 2005, 8, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Zhao, Y.; Xue, F.; Zheng, Y.; Huang, H.; Wang, W.; Chang, Y.; Yang, H.; Zhang, J. Exosomal DNA Aptamer Targeting α-Synuclein Aggregates Reduced Neuropathological Deficits in a Mouse Parkinson’s Disease Model. Mol. Ther.-Nucleic Acids 2019, 17, 726–740. [Google Scholar] [CrossRef] [Green Version]
- Izco, M.; Blesa, J.; Schleef, M.; Schmeer, M.; Porcari, R.; Al-Shawi, R.; Ellmerich, S.; de Toro, M.; Gardiner, C.; Seow, Y.; et al. Systemic Exosomal Delivery of shRNA Minicircles Prevents Parkinsonian Pathology. Mol. Ther. 2019, 27, 2111–2122. [Google Scholar] [CrossRef]
- Gao, X.; Ran, N.; Dong, X.; Zuo, B.; Yang, R.; Zhou, Q.; Moulton, H.M.; Seow, Y.; Yin, H.F. Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci. Transl. Med. 2018, 10, eaat0195. [Google Scholar] [CrossRef] [Green Version]
- Morgan, S.; Duguez, S.; Duddy, W. Personalized Medicine and Molecular Interaction Networks in Amyotrophic Lateral Sclerosis (ALS): Current Knowledge. J. Pers. Med. 2018, 8, 44. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.S.; Ahn, J.S.; Kim, S.; Kim, H.J.; Kim, S.H.; Kang, J.S. The potential theragnostic (diagnostic+therapeutic) application of exosomes in diverse biomedical fields. Korean J. Physiol. Pharmacol. 2018, 22, 113–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, M.; Liu, C.; Cook, T.J.; Bullock, K.M.; Zhao, Y.; Ginghina, C.; Li, Y.; Aro, P.; Dator, R.; He, C.; et al. Plasma exosomal α-synuclein is likely CNS-derived and increased in Parkinson’s disease. Acta Neuropathol. 2014, 128, 639–650. [Google Scholar] [CrossRef]
- Candelario, K.M.; Steindler, D.A. The role of extracellular vesicles in the progression of neurodegenerative disease and cancer. Trends Mol. Med. 2014, 20, 368–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boukouris, S.; Mathivanan, S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteom.-Clin. Appl. 2015, 9, 358–367. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Chen, K.; Wang, Z.; Wang, Y.; Liu, J.; Lin, L.; Shao, Y.; Gao, L.; Yin, H.; Cui, C.; et al. DNA in serum extracellular vesicles is stable under different storage conditions. BMC Cancer 2016, 16, 753. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, N.; Doi, H.; Kurata, Y.; Kagawa, H.; Atobe, Y.; Funakoshi, K.; Tada, M.; Katsumoto, A.; Tanaka, K.; Kunii, M.; et al. Proteomic analysis of exosome-enriched fractions derived from cerebrospinal fluid of amyotrophic lateral sclerosis patients. Neurosci. Res. 2019, 160, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Otake, K.; Kamiguchi, H.; Hirozane, Y. Identification of biomarkers for amyotrophic lateral sclerosis by comprehensive analysis of exosomal mRNAs in human cerebrospinal fluid. BMC Med. Genom. 2019, 12, 7. [Google Scholar] [CrossRef] [Green Version]
- Saucier, D.; Wajnberg, G.; Roy, J.; Beauregard, A.P.; Chacko, S.; Crapoulet, N.; Fournier, S.; Ghosh, A.; Lewis, S.M.; Marrero, A.; et al. Identification of a circulating miRNA signature in extracellular vesicles collected from amyotrophic lateral sclerosis patients. Brain Res. 2019, 1708, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xia, K.; Chen, L.; Fan, D. Increased Interleukin-6 Levels in the Astrocyte-Derived Exosomes of Sporadic Amyotrophic Lateral Sclerosis Patients. Front. Neurosci. 2019, 13, 574. [Google Scholar] [CrossRef] [Green Version]
- Katsu, M.; Hama, Y.; Utsumi, J.; Takashina, K.; Yasumatsu, H.; Mori, F.; Wakabayashi, K.; Shoji, M.; Sasaki, H. MicroRNA expression profiles of neuron-derived extracellular vesicles in plasma from patients with amyotrophic lateral sclerosis. Neurosci. Lett. 2019, 708, 134176. [Google Scholar] [CrossRef]
- Xu, Q.; Zhao, Y.; Zhou, X.; Luan, J.; Cui, Y.; Han, J. Comparison of the extraction and determination of serum exosome and miRNA in serum and the detection of miR-27a-3p in serum exosome of ALS patients. Intractable Rare Dis. Res. 2018, 7, 13–18. [Google Scholar] [CrossRef] [Green Version]
- Tao, Z.; Wang, H.; Xia, Q.; Li, K.; Li, K.; Jiang, X.; Xu, G.; Wang, G.; Ying, Z. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum. Mol. Genet. 2015, 24, 2426–2441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banack, S.A.; Dunlop, R.A.; Cox, P.A. An miRNA fingerprint using neural-enriched extracellular vesicles from blood plasma: Towards a biomarker for amyotrophic lateral sclerosis/motor neuron disease. Open Biol. 2020, 10, 200116. [Google Scholar] [CrossRef] [PubMed]


| Exosome Origin | ALIX | CD9 | CD63 | CD71 | CD81 | CD82 | Flotillin-1 | Hsp70 | Hsp90 | Lamp1 | Lamp2 | Rab7 | Rab11 | Tsg101 | Calnexin | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cortical neurons | +++ | + | +++ | ++ | [8,37,38,39] | |||||||||||
| Microglial cell | ++ | ++++++++ | +++++++ | + | +++ | + | +++ | + | + | + | + | [11,40,41,42,43,44,45,46,47,48,49,50] | ||||
| Oligodendrocytes | ++++++ | + | ++ | +++ | − − − − | [10,12,13,51,52,53] | ||||||||||
| Schwann cells | +++ | ++ | ++++ | + | ++++ | + | +++ | [54,55,56,57,58,59] | ||||||||
| Astrocytes | +++ | ++++ | + | +++ | ++ | + | ++++ | [9,60,61,62,63,64,65,66] | ||||||||
| Hippocampal neurons | + | + | [67] | |||||||||||||
| Motor neurons | + | + | + | [68] | ||||||||||||
| Skeletal muscle cells | +++++++++ | + | +++++ | + | +++++ | ++ | + | ++ | ++++++ | − − − | [69,70,71,72,73,74,75,76,77,78,79] |
| Exosome Origin | Ref | Exosome Marker | Density (g.mL−1) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.07 | 1.08 | 1.09 | 1.1 | 1.11 | 1.12 | 1.13 | 1.14 | 1.15 | 1.16 | 1.17 | 1.18 | 1.19 | 1.2 | |||
| Rat primary cortical neurons | [8] | ALIX | 100 | |||||||||||||
| Murine oligodendrocytes | [10] | 55 | ||||||||||||||
| Oligodendrocyte cell line | [12] | 95 | ||||||||||||||
| Oligodendrocyte cell line | [52] | 75 | ||||||||||||||
| Murine neuroglial cells | [124] | Tsg101 | 70 | |||||||||||||
| Oligodendrocyte cell line | [52] | 75 | ||||||||||||||
| Murine neuroglial cells | [124] | Hsc70/ Hsp70 | 70 | |||||||||||||
| Oligodendrocyte cell line | [12] | 95 | ||||||||||||||
| Murine neuroglial cells | [10] | Flotillin | 70 | |||||||||||||
| Oligodendrocyte cell line | [52] | |||||||||||||||
| Human embryonic myotubes | [72] | CD63 | 70 | |||||||||||||
| Human embryonic myotubes | [72] | CD81 | 70 | 70 | ||||||||||||
| Human embryonic myotubes | [72] | CD9 | 70 | 70 | ||||||||||||
| Human adult myotubes | [69] | CD63 | 100 | |||||||||||||
| Human adult myotubes | [69] | CD82 | 100 | |||||||||||||
| Exosome Ligand | Target Cell Ligand | Ref | |
|---|---|---|---|
| Glycoproteins | Fibronectin | Heparin sulfate proteoglycans (HSPGs) | [127] |
| Fibronectin | Integrins | [128] | |
| ICAM (CD54) | LFA-1 | [129,130,131] | |
| MUC1 | DC-SIGN | [132] | |
| Integrins | β1 and β2 integrins | ICAM-1 | [133] |
| β1 and β2 integrins | Collagen-I | ||
| β1 and β2 integrins | Fibronectin | ||
| Integrin α4β1 | Fibronectin | [134] | |
| αvβ3 / αvβ5 integrins | MFG-E8 | [135] | |
| CD47 | SIRPα | [136] | |
| Lectin | C-type lectin | Mannose-rich C-type lectin receptor | [137] |
| Galectin 5 | Glycoproteins (CD7, α5β1-integrin, or laminin) | [138] | |
| Galectin 9 | Tim 3 | [139] | |
| Tetraspanins | Tspan8-CD49d | ICAM-1 (CD54) | [140] |
| Lipid rafts | Phosphatidylserine | Tim-1/4 | [96] |
| Phosphatidylserine | MFG-E8 | [135] | |
| Phosphatidylethanolamine | MFG-E8 | ||
| Annexin 2 | Lipid raft domain | [141] | |
| Sugar | α2,3-linked sialic acids | sialoadhesin (CD169) | [142] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anakor, E.; Le Gall, L.; Dumonceaux, J.; Duddy, W.J.; Duguez, S. Exosomes in Ageing and Motor Neurone Disease: Biogenesis, Uptake Mechanisms, Modifications in Disease and Uses in the Development of Biomarkers and Therapeutics. Cells 2021, 10, 2930. https://doi.org/10.3390/cells10112930
Anakor E, Le Gall L, Dumonceaux J, Duddy WJ, Duguez S. Exosomes in Ageing and Motor Neurone Disease: Biogenesis, Uptake Mechanisms, Modifications in Disease and Uses in the Development of Biomarkers and Therapeutics. Cells. 2021; 10(11):2930. https://doi.org/10.3390/cells10112930
Chicago/Turabian StyleAnakor, Ekene, Laura Le Gall, Julie Dumonceaux, William John Duddy, and Stephanie Duguez. 2021. "Exosomes in Ageing and Motor Neurone Disease: Biogenesis, Uptake Mechanisms, Modifications in Disease and Uses in the Development of Biomarkers and Therapeutics" Cells 10, no. 11: 2930. https://doi.org/10.3390/cells10112930