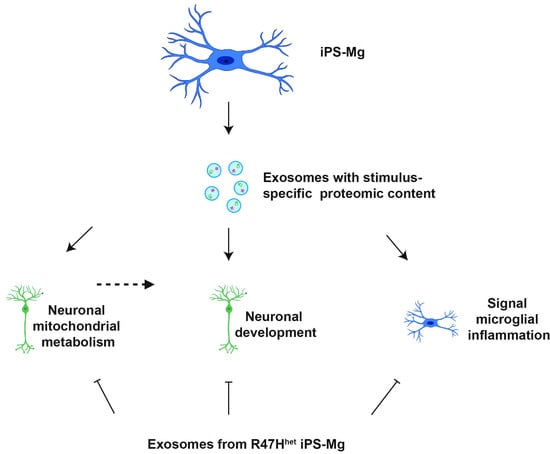
Differential Stimulation of Pluripotent Stem Cell-Derived Human Microglia Leads to Exosomal Proteomic Changes Affecting Neurons
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Cell Culture
2.2.1. iPS-Mg
2.2.2. iPS-Neurons
2.2.3. SH-SY5Y Neurons
2.3. Cell Treatment and Exosome Collection
2.4. Proteomic Analysis
2.5. Quantitative PCR
2.6. Immunocytochemistry
2.7. Metabolic Assays
2.8. Endotoxin Assay
2.9. Statistical Analysis
3. Results
3.1. Stimulatory Treatments Induce Specific Exosomal Proteomic Changes
3.2. R47Hhet Exosomes Contain More DAM-Associated Proteins
3.3. Network Analysis
3.4. Microglial Exosomes Can Transmit Inflammatory Messages
3.5. Basal Cv Exosomes Can Support Neuronal Development
3.6. Metabolic Effects of Exosomes on Neurons
4. Discussion
4.1. Influence of TREM2 on Exosomes from iPS-Mg
4.2. Stimulus Specificity Includes LPS-Specific Inflammatory Changes and PS+-Specific Metabolic Changes
4.3. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lambert, J.C.; Heath, S.; Even, G.; Campion, D.; Sleegers, K.; Hiltunen, M.; Combarros, O.; Zelenika, D.; Bullido, M.J.; Tavernier, B.; et al. Meta-Analysis of 74,046 Individuals Identifies 11 New Susceptibility Loci for Alzheimer’s Disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef] [Green Version]
- Guerreiro, R.; Wojtas, A.; Bras, J.; Carrasquillo, M.; Rogaeva, E.; Majounie, E.; Cruchaga, C.; Kauwe, J.S.K.; Younkin, S.; Hazrati, L.; et al. TREM-2 Variants in AD. N. Engl. J. Med. 2013, 368, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Reitz, C.; Jun, G.; Naj, A.; Rajbhandary, R.; Vardarajan, B.N.; Wang, L.S.; Valladares, O.; Lin, C.F.; Larson, E.B.; Graff-Radford, N.R.; et al. Variants in the ATP-Binding Cassette Transporter (ABCA7), Apolipoprotein e Ε4, and the Risk of Late-Onset Alzheimer Disease in African Americans. JAMA—J. Am. Med. Assoc. 2013, 309, 1483–1492. [Google Scholar] [CrossRef]
- Gjoneska, E.; Pfenning, A.R.; Mathys, H.; Quon, G.; Kundaje, A.; Tsai, L.H.; Kellis, M. Conserved Epigenomic Signals in Mice and Humans Reveal Immune Basis of Alzheimer’s Disease. Nature 2015, 518, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Daws, M.R.; Sullam, P.M.; Niemi, E.C.; Chen, T.T.; Tchao, N.K.; Seaman, W.E. Pattern Recognition by TREM-2: Binding of Anionic Ligands. J. Immunol. 2003, 171, 594–599. [Google Scholar] [CrossRef] [Green Version]
- Filipello, F.; Morini, R.; Corradini, I.; Zerbi, V.; Canzi, A.; Michalski, B.; Erreni, M.; Markicevic, M.; Starvaggi-Cucuzza, C.; Otero, K.; et al. The Microglial Innate Immune Receptor TREM2 Is Required for Synapse Elimination and Normal Brain Connectivity. Immunity 2018, 48, 979–991.e8. [Google Scholar] [CrossRef] [Green Version]
- Shirotani, K.; Hori, Y.; Yoshizaki, R.; Higuchi, E.; Colonna, M.; Saito, T.; Hashimoto, S.; Saito, T.; Saido, T.C.; Iwata, N.; et al. Aminophospholipids Are Signal-Transducing TREM2 Ligands on Apoptotic Cells. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Jonsson, T.; Stefansson, H.; Steinberg, S.; Jonsdottir, I.; Jonsson, P.V.; Snaedal, J.; Bjornsson, S.; Huttenlocher, J.; Levey, A.I.; Lah, J.J.; et al. Variant of TREM2 Associated with the Risk of Alzheimer’s Disease. N. Engl. J. Med. 2013, 368, 107–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Reitboeck, P.; Phillips, A.; Piers, T.M.; Villegas-Llerena, C.; Butler, M.; Mallach, A.; Rodrigues, C.; Arber, C.E.; Heslegrave, A.; Zetterberg, H.; et al. Human Induced Pluripotent Stem Cell-Derived Microglia-Like Cells Harboring TREM2 Missense Mutations Show Specific Deficits in Phagocytosis. Cell Rep. 2018, 24, 2300–2311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piers, T.M.; Cosker, K.; Mallach, A.; Johnson, G.T.; Guerreiro, R.; Hardy, J.; Pocock, J.M. A Locked Immunometabolic Switch Underlies TREM2 R47H Loss of Function in Human IPSC-Derived Microglia. Faseb J. 2020, 34, 2436–2450. [Google Scholar] [CrossRef] [Green Version]
- Ulland, T.K.; Song, W.M.; Huang, S.C.C.; Ulrich, J.D.; Sergushichev, A.; Beatty, W.L.; Loboda, A.A.; Zhou, Y.; Cairns, N.J.; Kambal, A.; et al. TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell 2017, 170, 649–663.e13. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Zhang, Z.L.; Li, X.; Liao, C.; Mou, P.; Wang, T.; Wang, Z.; Wang, Z.; Wei, M.; Xu, H.; et al. TREM2/DAP12 Complex Regulates Inflammatory Responses in Microglia via the JNK Signaling Pathway. Front. Aging Neurosci. 2017, 9, 1–9. [Google Scholar] [CrossRef]
- Hooper, C.; Sainz-Fuertes, R.; Lynham, S.; Hye, A.; Killick, R.; Warley, A.; Bolondi, C.; Pocock, J.; Lovestone, S. Wnt3a Induces Exosome Secretion from Primary Cultured Rat Microglia. Bmc Neurosci. 2012, 13, 144. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Boza-Serrano, A.; Dunning, C.J.R.; Clausen, B.H.; Lambertsen, K.L.; Deierborg, T. Inflammation Leads to Distinct Populations of Extracellular Vesicles from Microglia. J. Neuroinflamm. 2018, 15, 168. [Google Scholar] [CrossRef]
- Tamboli, I.Y.; Barth, E.; Christian, L.; Siepmann, M.; Kumar, S.; Singh, S.; Tolksdorf, K.; Heneka, M.T.; Lütjohann, D.; Wunderlich, P.; et al. Statins Promote the Degradation of Extracellular Amyloid β-Peptide by Microglia via Stimulation of Exosome-Associated Insulin-Degrading Enzyme (IDE) Secretion. J. Biol. Chem. 2010, 285, 37405–37414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuyama, K.; Sun, H.; Mitsutake, S.; Igarashi, Y. Sphingolipid-Modulated Exosome Secretion Promotes Clearance of Amyloid-β by Microglia. J. Biol. Chem. 2012, 287, 10977–10989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, P.; Turola, E.; Ruiz, A.; Bergami, A.; Libera, D.D.; Benussi, L.; Giussani, P.; Magnani, G.; Comi, G.; Legname, G.; et al. Microglia Convert Aggregated Amyloid-β into Neurotoxic Forms through the Shedding of Microvesicles. Cell Death Differ. 2014, 21, 582–593. [Google Scholar] [CrossRef] [Green Version]
- Asai, H.; Ikezu, S.; Tsunoda, S.; Medalla, M.; Luebke, J.; Haydar, T.; Wolozin, B.; Butovsky, O.; Kügler, S.; Ikezu, T. Depletion of Microglia and Inhibition of Exosome Synthesis Halt Tau Propagation. Nat. Neurosci. 2015, 18, 1584–1593. [Google Scholar] [CrossRef]
- Antonucci, F.; Turola, E.; Riganti, L.; Caleo, M.; Gabrielli, M.; Perrotta, C.; Novellino, L.; Clementi, E.; Giussani, P.; Viani, P.; et al. Microvesicles Released from Microglia Stimulate Synaptic Activity via Enhanced Sphingolipid Metabolism. Embo J. 2012, 31, 1231–1240. [Google Scholar] [CrossRef]
- Zhao, C.; Deng, Y.; He, Y.; Huang, X.; Wang, C.; Li, W. Decreased Level of Exosomal MiR-5121 Released from Microglia Suppresses Neurite Outgrowth and Synapse Recovery of Neurons Following Traumatic Brain Injury. Neurotherapeutics 2021, 18, 1273–1294. [Google Scholar] [CrossRef]
- Mallach, A.; Gobom, J.; Zetterberg, H.; Hardy, J.; Piers, T.M.; Wray, S.; Pocock, J.M. The Influence of the R47H TREM2 Variant on Microglial Exosome Profiles. Brain Commun. 2021, 3, fcab009. [Google Scholar] [CrossRef]
- Xiang, X.; Piers, T.M.; Wefers, B.; Zhu, K.; Mallach, A.; Brunner, B.; Kleinberger, G.; Song, W.; Colonna, M.; Herms, J.; et al. The Trem2 R47H Alzheimer’s Risk Variant Impairs Splicing and Reduces Trem2 MRNA and Protein in Mice but Not in Humans. Mol. Neurodegener. 2018, 13, 1–14. [Google Scholar] [CrossRef] [Green Version]
- van Wilgenburg, B.; Browne, C.; Vowles, J.; Cowley, S.A. Efficient, Long Term Production of Monocyte-Derived Macrophages from Human Pluripotent Stem Cells under Partly-Defined and Fully-Defined Conditions. PLos ONE 2013, 8, e71098. [Google Scholar] [CrossRef]
- Cosker, K.; Mallach, A.; Limaye, J.; Piers, T.M.; Staddon, J.; Neame, S.; Hardy, J.; Pocock, J.M. Microglial Signalling Pathway Deficits Associated with the Patient Derived R47H TREM2 Variants Linked to AD Indicate Inability to Activate Inflammasome. Sci. Rep. 2021, 11, 13316. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Kirwan, P.; Livesey, F.J. Directed Differentiation of Human Pluripotent Stem Cells to Cerebral Cortex Neurons and Neural Networks. Nat. Protoc. 2012, 7, 1836–1846. [Google Scholar] [CrossRef] [PubMed]
- Arber, C.; Toombs, J.; Lovejoy, C.; Ryan, N.S.; Paterson, R.W.; Willumsen, N.; Gkanatsiou, E.; Portelius, E.; Blennow, K.; Heslegrave, A.; et al. Familial Alzheimer’s Disease Patient-Derived Neurons Reveal Distinct Mutation-Specific Effects on Amyloid Beta. Mol. Psychiatry 2020, 25, 2919–2931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Encinas, M.; Iglesias, M.; Liu, Y.; Wang, H.; Muhaisen, A.; Cena, V.; Gallego, C.; Comella, J.X. Sequential Treatment of SH-SY5Y Cells with Retinoic Acid and Brain-Derived Neurotrophic Factor Gives Rise to Fully Differentiated, Neutrophic Factor-Dependent, Human Neuron-Like Cells. J. Neurochem. 2000, 75, 991–1003. [Google Scholar] [CrossRef]
- Forster, J.I.; Köglsberger, S.; Trefois, C.; Boyd, O.; Baumuratov, A.S.; Buck, L.; Balling, R.; Antony, P.M.A. Characterization of Differentiated SH-SY5Y as Neuronal Screening Model Reveals Increased Oxidative Vulnerability. J. Biomol. Screen. 2015, 21, 496–509. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, C.L.; Koike, M.; Spusta, S.C.; Niemi, E.C.; Yenari, M.; Nakamura, M.C.; Seaman, W.E. A Role for TREM2 Ligands in the Phagocytosis of Apoptotic Neuronal Cells by Microglia. J. Neurochem. 2009, 109, 1144–1156. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. Bmc Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Luo, R.; Oldham, M.C.; Horvath, S. Is My Network Module Preserved and Reproducible? Plos Comput. Biol. 2011, 7, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbajosa, G.; Malki, K.; Lawless, N.; Wang, H.; Ryder, J.W.; Wozniak, E.; Wood, K.; Mein, C.A.; Dobson, R.J.B.; Collier, D.A.; et al. Loss of Trem2 in Microglia Leads to Widespread Disruption of Cell Coexpression Networks in Mouse Brain. Neurobiol. Aging 2018, 69, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Young, K.; Morrison, H. Quantifying Microglia Morphology from Photomicrographs of Immunohistochemistry Prepared Tissue Using Imagej. J. Vis. Exp. 2018, 2018, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallach, A.; Weinert, M.; Arthur, J.; Gveric, D.; Tierney, T.S.; Alavian, K.N. Post Mortem Examination of Parkinson’s Disease Brains Suggests Decline in Mitochondrial Biomass, Reversed by Deep Brain Stimulation of Subthalamic Nucleus. Faseb J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2019, 33, 6957–6961. [Google Scholar] [CrossRef]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e17. [Google Scholar] [CrossRef] [PubMed]
- Butovsky, O.; Weiner, H.L. Microglial Signatures and Their Role in Health and Disease. Nat. Rev. Neurosci. 2018, 19, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Rangaraju, S.; Dammer, E.B.; Raza, S.A.; Rathakrishnan, P.; Xiao, H.; Gao, T.; Duong, D.M.; Pennington, M.W.; Lah, J.J.; Seyfried, N.T.; et al. Identification and Therapeutic Modulation of a Pro-Inflammatory Subset of Disease-Associated-Microglia in Alzheimer’s Disease. Mol. Neurodegener. 2018, 13, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, D.L.; Baratt, A.; Baric, R.S.; Kawaoka, Y.; Smith, R.D.; Orwoll, E.S.; Katze, M.G.; McWeeney, S.K. Protein Co-Expression Network Analysis (ProCoNA). J. Clin. Bioinform. 2013, 3, 1. [Google Scholar] [CrossRef] [Green Version]
- Morgan, S.C.; Taylor, D.L.; Pocock, J.M. Microglia Release Activators of Neuronal Proliferation Mediated by Activation of Mitogen-Activated Protein Kinase, Phosphatidylinositol-3-Kinase/Akt and Delta-Notch Signalling Cascades. J. Neurochem. 2004, 90, 89–101. [Google Scholar] [CrossRef]
- London, A.; Cohen, M.; Schwartz, M. Microglia and Monocyte-Derived Macrophages: Functionally Distinct Populations That Act in Concert in CNS Plasticity and Repair. Front. Cell. Neurosci. 2013, 7, 34. [Google Scholar] [CrossRef] [Green Version]
- Campos, P.B.; Paulsen, B.S.; Rehen, S.K. Accelerating Neuronal Aging in in Vitro Model Brain Disorders: A Focus on Reactive Oxygen Species. Front. Aging Neurosci. 2014, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Meiri, K.F.; Saffell, J.L.; Walsh, F.S.; Doherty, P. Neurite Outgrowth Stimulated by Neural Cell Adhesion Molecules Requires Growth-Associated Protein-43 (GAP-43) Function and Is Associated with GAP-43 Phosphorylation in Growth Cones. J. Neurosci. 1998, 18, 10429–10437. [Google Scholar] [CrossRef] [PubMed]
- Korshunova, I.; Novitskaya, V.; Kiryushko, D.; Pedersen, N.; Kolkova, K.; Kropotova, E.; Mosevitsky, M.; Rayko, M.; Morrow, J.S.; Ginzburg, I.; et al. GAP-43 Regulates NCAM-180-Mediated Neurite Outgrowth. J. Neurochem. 2007, 100, 1599–1612. [Google Scholar] [CrossRef]
- Cheng, A.; Wan, R.; Yang, J.-L.; Kamimura, N.; Son, T.G.; Ouyang, X.; Luo, Y.; Okun, E.; Mattson, M.P. Involvement of PGC-1α in the Formation and Maintenance of Neuronal Dendritic Spines. Nat. Commun. 2012, 3, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onishi, Y.; Ueha, T.; Kawamoto, T.; Hara, H.; Toda, M.; Harada, R.; Minoda, M.; Kurosaka, M.; Akisue, T. Regulation of Mitochondrial Proliferation by PGC-1α Induces Cellular Apoptosis in Musculoskeletal Malignancies. Sci. Rep. 2014, 4, 4–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh, J.I.; Kawana, N.; Yamamoto, Y. Pathway Analysis of ChIP-Seq-Based NRF1 Target Genes Suggests a Logical Hypothesis of Their Involvement in the Pathogenesis of Neurodegenerative Diseases. Gene Regul. Syst. Biol. 2013, 2013, 139–152. [Google Scholar] [CrossRef] [Green Version]
- Kiyama, T.; Chen, C.-K.; Wang, S.W.; Pan, P.; Ju, Z.; Wang, J.; Takada, S.; Klein, W.H.; Mao, C.-A. Essential Roles of Mitochondrial Biogenesis Regulator Nrf1 in Retinal Development and Homeostasis. Mol. Neurodegener. 2018, 13, 1–23. [Google Scholar] [CrossRef]
- Veereshwarayya, V.; Kumar, P.; Rosen, K.M.; Mestril, R.; Querfurth, H.W. Differential Effects of Mitochondrial Heat Shock Protein 60 and Related Molecular Chaperones to Prevent Intracellular β-Amyloid-Induced Inhibition of Complex IV and Limit Apoptosis. J. Biol. Chem. 2006, 281, 29468–29478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böttinger, L.; Oeljeklaus, S.; Guiard, B.; Rospert, S.; Warscheid, B.; Becker, T. Mitochondrial Heat Shock Protein (Hsp) 70 and Hsp10 Cooperate in the Formation of Hsp60 Complexes. J. Biol. Chem. 2015, 290, 11611–11622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, J.P.; Wardelmann, K.; Grune, T.; Kleinridders, A. Mitochondrial Chaperones in the Brain: Safeguarding Brain Health and Metabolism? Front. Endocrinol. 2018, 9, 196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, A.; Wake, H.; Ishikawa, A.W.; Eto, K.; Shibata, K.; Murakoshi, H.; Koizumi, S.; Moorhouse, A.J.; Yoshimura, Y.; Nabekura, J. Microglia Contact Induces Synapse Formation in Developing Somatosensory Cortex. Nat. Commun. 2016, 7, 12540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamberlain, K.A.; Sheng, Z.H. Mechanisms for the Maintenance and Regulation of Axonal Energy Supply. J. Neurosci. Res. 2019, 97, 897–913. [Google Scholar] [CrossRef] [Green Version]
- Kiryu-Seo, S.; Kiyama, H. Mitochondrial Behavior during Axon Regeneration/Degeneration in Vivo. Neurosci. Res. 2019, 139, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Parkhurst, C.N.; Yang, G.; Ninan, I.; Savas, J.N.; Yates, J.R.; Lafaille, J.J.; Hempstead, B.L.; Littman, D.R.; Gan, W.-B. Microglia Promote Learning-Dependent Synapse Formation through Brain-Derived Neurotrophic Factor. Cell 2013, 155, 1596–1609. [Google Scholar] [CrossRef] [Green Version]
- Crotti, A.; Ransohoff, R.M. Microglial Physiology and Pathophysiology: Insights from Genome-Wide Transcriptional Profiling. Immunity 2016, 44, 505–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamerman, J.A.; Jarjoura, J.R.; Humphrey, M.B.; Nakamura, M.C.; Seaman, W.E.; Lanier, L.L. Cutting Edge: Inhibition of TLR and FcR Responses in Macrophages by Triggering Receptor Expressed on Myeloid Cells (TREM)-2 and DAP12. J. Immunol. 2006, 177, 2051–2055. [Google Scholar] [CrossRef]
- Turnbull, I.R.; Gilfillan, S.; Cella, M.; Aoshi, T.; Miller, M.; Piccio, L.; Hernandez, M.; Colonna, M. Cutting Edge: TREM-2 Attenuates Macrophage Activation. J. Immunol. 2006, 177, 3520–3524. [Google Scholar] [CrossRef]
- Liu, T.; Zhu, B.; Liu, Y.; Zhang, X.; Yin, J.; Li, X.; Jiang, L.; Hodges, A.P.; Rosenthal, S.B.; Zhou, L.; et al. Multi-Omic Comparison of Alzheimer’s Variants in Human ESC-Derived Microglia Reveals Convergence at APOE. J. Exp. Med. 2020, 217, 12. [Google Scholar] [CrossRef]
- Sousa, C.; Golebiewska, A.; Poovathingal, S.K.; Kaoma, T.; Pires-Afonso, Y.; Martina, S.; Coowar, D.; Azuaje, F.; Skupin, A.; Balling, R.; et al. Single-cell Transcriptomics Reveals Distinct Inflammation-induced Microglia Signatures. Embo Rep. 2018, 19, 1–17. [Google Scholar] [CrossRef]
- Choi, D.S.; Kim, D.K.; Kim, Y.K.; Gho, Y.S. Proteomics, Transcriptomics and Lipidomics of Exosomes and Ectosomes. Proteomics 2013, 13, 1554–1571. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, W.; Freeman, M.L.; Lederman, M.M.; Vasilieva, E.; Romero, R.; Margolis, L. A System of Cytokines Encapsulated in ExtraCellular Vesicles. Sci. Rep. 2018, 8, 8973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puhka, M.; Takatalo, M.; Nordberg, M.E.; Valkonen, S.; Nandania, J.; Aatonen, M.; Yliperttula, M.; Laitinen, S.; Velagapudi, V.; Mirtti, T.; et al. Metabolomic Profiling of Extracellular Vesicles and Alternative Normalization Methods Reveal Enriched Metabolites and Strategies to Study Prostate Cancer-Related Changes. Theranostics 2017, 7, 3824–3841. [Google Scholar] [CrossRef] [PubMed]
- Trotta, T.; Panaro, M.A.; Cianciulli, A.; Mori, G.; Di Benedetto, A.; Porro, C. Microglia-Derived Extracellular Vesicles in Alzheimer’s Disease: A Double-Edged Sword. Biochem. Pharm. 2018. [Google Scholar] [CrossRef] [PubMed]
- Potolicchio, I.; Carven, G.J.; Xu, X.; Stipp, C.; Riese, R.J.; Stern, L.J.; Santambrogio, L. Proteomic Analysis of Microglia-Derived Exosomes: Metabolic Role of the Aminopeptidase CD13 in Neuropeptide Catabolism. J. Immunol. 2005, 175, 2237–2243. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Gene Name | Identifier |
|---|---|
| APOE | Hs00171168_m1 |
| AXL | Hs01064444_m1 |
| CSF1R | Hs00911250_m1 |
| GAPDH | Hs02758991_g1 |
| GPNMB | Hs01095669_m1 |
| GRN | Hs00963707_g1 |
| HSPD1 | Hs01036753_g1 |
| IL1B | Hs00174097_m1 |
| IL6 | Hs00985639_m1 |
| PPARGC | Hs01016719_m1 |
| TNF | Hs01113624_g1 |
| TREM2 | Hs00219132_m1 |
| TYROBP | Hs00182426_m1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mallach, A.; Gobom, J.; Arber, C.; Piers, T.M.; Hardy, J.; Wray, S.; Zetterberg, H.; Pocock, J. Differential Stimulation of Pluripotent Stem Cell-Derived Human Microglia Leads to Exosomal Proteomic Changes Affecting Neurons. Cells 2021, 10, 2866. https://doi.org/10.3390/cells10112866
Mallach A, Gobom J, Arber C, Piers TM, Hardy J, Wray S, Zetterberg H, Pocock J. Differential Stimulation of Pluripotent Stem Cell-Derived Human Microglia Leads to Exosomal Proteomic Changes Affecting Neurons. Cells. 2021; 10(11):2866. https://doi.org/10.3390/cells10112866
Chicago/Turabian StyleMallach, Anna, Johan Gobom, Charles Arber, Thomas M. Piers, John Hardy, Selina Wray, Henrik Zetterberg, and Jennifer Pocock. 2021. "Differential Stimulation of Pluripotent Stem Cell-Derived Human Microglia Leads to Exosomal Proteomic Changes Affecting Neurons" Cells 10, no. 11: 2866. https://doi.org/10.3390/cells10112866