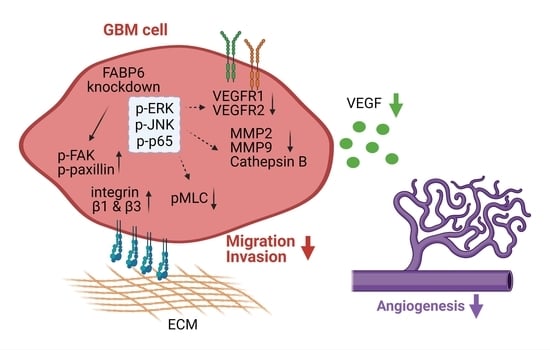
Inhibition of FABP6 Reduces Tumor Cell Invasion and Angiogenesis through the Decrease in MMP-2 and VEGF in Human Glioblastoma Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Immunohistochemical Staining of Human Glioma Specimens
2.2. Cell Culture
2.3. Antibodies
2.4. Stable Expression of shRNAs
2.5. Real-Time Polymerase Chain Reaction (RT-PCR)
2.6. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Assay
2.7. Cell Migration and Invasion Assays
2.8. Western Blotting
2.9. Tube Formation Assay
2.10. Orthotopic Xenograft Animal Model
2.11. Histological and Immunohistochemical Examination
2.12. Terminal Deoxynucleotidyl Transferase (TdT) dUTP Nick-End Labeling (TUNEL) Assay
2.13. Enzyme-Linked Immunosorbent Assay (ELISA) for VEGF in Condition Medium
2.14. Statistical Analysis
3. Results
3.1. FABP6 Had Higher Expression in Glioma
3.2. Analysis of the Expression of FABP and Establishment of shRNA (shFABP6) Stable Clones in Glioma Cell Lines
3.3. Knockdown of FABP6 Decreased the Migration and Invasion Abilities of Invasion-Related Proteins in the LN229 and U87MG Cells
3.4. Tube Formation of Endothelial Cells Was Attenuated by the Knockdown of FABP6 in Glioma Cells
3.5. The Reduced Phosphorylation of JNK, ERK, and p65 Was Caused by FABP6 Inhibition
3.6. FABP6 Knockdown Combined with TMZ Application Attenuated Tumor Progression in the Orthotropic Xenograft Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro-Oncology 2014, 16, 896–913. [Google Scholar] [CrossRef] [Green Version]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Marsden, P.A. Angiogenesis in glioblastoma. N. Engl. J. Med. 2013, 369, 1561–1563. [Google Scholar] [CrossRef] [Green Version]
- Jing, C.; Beesley, C.; Foster, C.S.; Rudland, P.S.; Fujii, H.; Ono, T.; Chen, H.; Smith, P.H.; Ke, Y. Identification of the messenger RNA for human cutaneous fatty acid-binding protein as a metastasis inducer. Cancer Res. 2000, 60, 2390–2398. [Google Scholar]
- Tolle, A.; Suhail, S.; Jung, M.; Jung, K.; Stephan, C. Fatty acid binding proteins (FABPs) in prostate, bladder and kidney cancer cell lines and the use of IL-FABP as survival predictor in patients with renal cell carcinoma. BMC Cancer 2011, 11, 302. [Google Scholar] [CrossRef] [Green Version]
- Amiri, M.; Yousefnia, S.; Seyed Forootan, F.; Peymani, M.; Ghaedi, K.; Nasr Esfahani, M.H. Diverse roles of fatty acid binding proteins (FABPs) in development and pathogenesis of cancers. Gene 2018, 676, 171–183. [Google Scholar] [CrossRef]
- Huang, M.; Narita, S.; Inoue, T.; Koizumi, A.; Saito, M.; Tsuruta, H.; Numakura, K.; Satoh, S.; Nanjo, H.; Sasaki, T.; et al. Fatty acid binding protein 4 enhances prostate cancer progression by upregulating matrix metalloproteinases and stromal cell cytokine production. Oncotarget 2017, 8, 111780–111794. [Google Scholar] [CrossRef] [Green Version]
- Elmasri, H.; Ghelfi, E.; Yu, C.W.; Traphagen, S.; Cernadas, M.; Cao, H.; Shi, G.P.; Plutzky, J.; Sahin, M.; Hotamisligil, G.; et al. Endothelial cell-fatty acid binding protein 4 promotes angiogenesis: Role of stem cell factor/c-kit pathway. Angiogenesis 2012, 15, 457–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohmachi, T.; Inoue, H.; Mimori, K.; Tanaka, F.; Sasaki, A.; Kanda, T.; Fujii, H.; Yanaga, K.; Mori, M. Fatty acid binding protein 6 is overexpressed in colorectal cancer. Clin. Cancer Res. 2006, 12, 5090–5095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rosa, A.; Pellegatta, S.; Rossi, M.; Tunici, P.; Magnoni, L.; Speranza, M.C.; Malusa, F.; Miragliotta, V.; Mori, E.; Finocchiaro, G.; et al. A radial glia gene marker, fatty acid binding protein 7 (FABP7), is involved in proliferation and invasion of glioblastoma cells. PLoS ONE 2012, 7, e52113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, D.D.; Robbins, M.E.; Spector, A.A.; Wen, B.C.; Hussey, D.H. The fatty acid composition of human gliomas differs from that found in nonmalignant brain tissue. Lipids 1996, 31, 1283–1288. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta 2007, 1773, 1263–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Wang, H.; Zhang, W.; Huang, H.J.; Liao, W.S.; Fuller, G.N. Analysis of the activation status of Akt, NFkappaB, and Stat3 in human diffuse gliomas. Lab. Investig. 2004, 84, 941–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puliyappadamba, V.T.; Hatanpaa, K.J.; Chakraborty, S.; Habib, A.A. The role of NF-kappaB in the pathogenesis of glioma. Mol. Cell Oncol. 2014, 1, e963478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhao, X.; Deng, L.; Li, X.; Wang, G.; Li, Y.; Chen, M. High expression of FABP4 and FABP6 in patients with colorectal cancer. World J. Surg. Oncol. 2019, 17, 171. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Sun, Q.; Patel, D.; Stommel, J.M. Cholesterol metabolism: A potential therapeutic target in glioblastoma. Cancers 2019, 11, 146. [Google Scholar] [CrossRef] [Green Version]
- Elsherbiny, M.E.; Emara, M.; Godbout, R. Interaction of brain fatty acid-binding protein with the polyunsaturated fatty acid environment as a potential determinant of poor prognosis in malignant glioma. Prog. Lipid Res. 2013, 52, 562–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Totsukawa, G.; Wu, Y.; Sasaki, Y.; Hartshorne, D.J.; Yamakita, Y.; Yamashiro, S.; Matsumura, F. Distinct roles of MLCK and ROCK in the regulation of membrane protrusions and focal adhesion dynamics during cell migration of fibroblasts. J. Cell Biol. 2004, 164, 427–439. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Liu, Y.; You, J.; Zhang, H.; Zhang, X.; Ye, L. Myosin light-chain kinase contributes to the proliferation and migration of breast cancer cells through cross-talk with activated ERK1/2. Cancer Lett. 2008, 270, 312–327. [Google Scholar] [CrossRef]
- Lopez-Colome, A.M.; Lee-Rivera, I.; Benavides-Hidalgo, R.; Lopez, E. Paxillin: A crossroad in pathological cell migration. J. Hematol. Oncol. 2017, 10, 50. [Google Scholar] [CrossRef] [Green Version]
- Yano, H.; Uchida, H.; Iwasaki, T.; Mukai, M.; Akedo, H.; Nakamura, K.; Hashimoto, S.; Sabe, H. Paxillin alpha and Crk-associated substrate exert opposing effects on cell migration and contact inhibition of growth through tyrosine phosphorylation. Proc. Natl. Acad. Sci. USA 2000, 97, 9076–9081. [Google Scholar] [CrossRef] [Green Version]
- Deryugina, E.I.; Quigley, J.P. Tumor angiogenesis: MMP-mediated induction of intravasation- and metastasis-sustaining neovasculature. Matrix. Biol. 2015, 44–46, 94–112. [Google Scholar] [CrossRef]
- Fang, L.Y.; Wong, T.Y.; Chiang, W.F.; Chen, Y.L. Fatty-acid-binding protein 5 promotes cell proliferation and invasion in oral squamous cell carcinoma. J. Oral. Pathol. Med. 2010, 39, 342–348. [Google Scholar] [CrossRef]
- Liao, B.; Geng, L.; Zhang, F.; Shu, L.; Wei, L.; Yeung, P.K.K.; Lam, K.S.L.; Chung, S.K.; Chang, J.; Vanhoutte, P.M.; et al. Adipocyte fatty acid-binding protein exacerbates cerebral ischaemia injury by disrupting the blood-brain barrier. Eur. Heart J. 2020, 41, 3169–3180. [Google Scholar] [CrossRef]
- Tian, W.; Zhang, W.; Zhang, Y.; Zhu, T.; Hua, Y.; Li, H.; Zhang, Q.; Xia, M. FABP4 promotes invasion and metastasis of colon cancer by regulating fatty acid transport. Cancer Cell Int. 2020, 20, 512. [Google Scholar] [CrossRef]
- Hamer, I.; Delaive, E.; Dieu, M.; Abdel-Sater, F.; Mercy, L.; Jadot, M.; Arnould, T. Up-regulation of cathepsin B expression and enhanced secretion in mitochondrial DNA-depleted osteosarcoma cells. Biol. Cell 2009, 101, 31–41. [Google Scholar] [CrossRef]
- Perazzoli, G.; Prados, J.; Ortiz, R.; Caba, O.; Cabeza, L.; Berdasco, M.; Gonzalez, B.; Melguizo, C. Temozolomide Resistance in Glioblastoma Cell Lines: Implication of MGMT, MMR, P-Glycoprotein and CD133 Expression. PLoS ONE 2015, 10, e0140131. [Google Scholar] [CrossRef] [Green Version]
- Hermisson, M.; Klumpp, A.; Wick, W.; Wischhusen, J.; Nagel, G.; Roos, W.; Kaina, B.; Weller, M. O6-methylguanine DNA methyltransferase and p53 status predict temozolomide sensitivity in human malignant glioma cells. J. Neurochem. 2006, 96, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuhashi, M.; Tuncman, G.; Görgün, C.Z.; Makowski, L.; Atsumi, G.; Vaillancourt, E.; Kono, K.; Babaev, V.R.; Fazio, S.; Linton, M.F. Treatment of diabetes and atherosclerosis by inhibiting fatty-acid-binding protein aP2. Nature 2007, 447, 959–965. [Google Scholar] [CrossRef] [Green Version]
- Sulsky, R.; Magnin, D.R.; Huang, Y.; Simpkins, L.; Taunk, P.; Patel, M.; Zhu, Y.; Stouch, T.R.; Bassolino-Klimas, D.; Parker, R. Potent and selective biphenyl azole inhibitors of adipocyte fatty acid binding protein (aFABP). Bioorganic Med. Chem. Lett. 2007, 17, 3511–3515. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Xiao, H.; Liao, R.; Chen, Q.; Peng, C.; Zhang, Y.; Mu, T.; Wu, Z. Fatty acid binding protein 5 promotes tumor angiogenesis and activates the IL6/STAT3/VEGFA pathway in hepatocellular carcinoma. Biomed. Pharm. 2018, 106, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Ku, C.Y.; Liu, Y.H.; Lin, H.Y.; Lu, S.C.; Lin, J.Y. Liver fatty acid-binding protein (L-FABP) promotes cellular angiogenesis and migration in hepatocellular carcinoma. Oncotarget 2016, 7, 18229–18246. [Google Scholar] [CrossRef] [Green Version]
- Harjes, U.; Bridges, E.; Gharpure, K.M.; Roxanis, I.; Sheldon, H.; Miranda, F.; Mangala, L.S.; Pradeep, S.; Lopez-Berestein, G.; Ahmed, A.; et al. Antiangiogenic and tumour inhibitory effects of downregulating tumour endothelial FABP4. Oncogene 2017, 36, 912–921. [Google Scholar] [CrossRef] [Green Version]







| Name | Species | Brand | Catalog Number |
|---|---|---|---|
| Cathepsin B | Rb | Abcam | ab125067 |
| CD31 | Rb | Abcam | ab28364 |
| ERK | Rb | Cell Signaling Technology | 9102s |
| FABP6 | Rb | Novus | NBP1-32482 |
| FAK | Rb | Cell Signaling Technology | 71433s |
| GAPDH | Rb | Cell Signaling Technology | 5174s |
| JNK | Rb | Cell Signaling Technology | 9252s |
| MLC 2 | Rb | Cell Signaling Technology | 8505s |
| MMP2 | Rb | Cell Signaling Technology | 13132s |
| MMP9 | Rb | Cell Signaling Technology | 13667s |
| P65 | Rb | Cell Signaling Technology | 8242s |
| Paxillin | Ms | BD Biosciences | 610051 |
| p-ERK | Rb | Cell Signaling Technology | 4377s |
| p-FAK (Y397) | Ms | BD Biosciences | 611723 |
| p-JNK | Rb | Cell Signaling Technology | 4668s |
| p-MLC (Ser19) | Rb | Cell Signaling Technology | 3671 |
| p-p65 | Rb | Abcam | Ab185619 |
| p-paxillin | Rb | Cell Signaling Technology | 2541s |
| TIMP-1 | Rb | Abcam | ab109125 |
| TIMP-2 | Ms | Millipore | MAB13446 |
| VEGFR1 | Rb | Abcam | ab32152 |
| VEGFR2 | Rb | Cell Signaling Technology | 2479s |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pai, F.-C.; Huang, H.-W.; Tsai, Y.-L.; Tsai, W.-C.; Cheng, Y.-C.; Chang, H.-H.; Chen, Y. Inhibition of FABP6 Reduces Tumor Cell Invasion and Angiogenesis through the Decrease in MMP-2 and VEGF in Human Glioblastoma Cells. Cells 2021, 10, 2782. https://doi.org/10.3390/cells10102782
Pai F-C, Huang H-W, Tsai Y-L, Tsai W-C, Cheng Y-C, Chang H-H, Chen Y. Inhibition of FABP6 Reduces Tumor Cell Invasion and Angiogenesis through the Decrease in MMP-2 and VEGF in Human Glioblastoma Cells. Cells. 2021; 10(10):2782. https://doi.org/10.3390/cells10102782
Chicago/Turabian StylePai, Feng-Cheng, Hsiang-Wei Huang, Yu-Ling Tsai, Wen-Chiuan Tsai, Yu-Chen Cheng, Hsin-Han Chang, and Ying Chen. 2021. "Inhibition of FABP6 Reduces Tumor Cell Invasion and Angiogenesis through the Decrease in MMP-2 and VEGF in Human Glioblastoma Cells" Cells 10, no. 10: 2782. https://doi.org/10.3390/cells10102782