Improved Denitrification Performance of Polybutylene Succinate/Corncob Composite Carbon Source by Proper Pretreatment: Performance, Functional Genes and Microbial Community Structure
Abstract
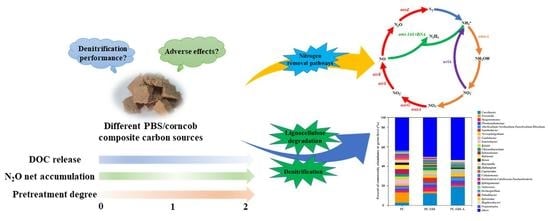
:1. Introduction
2. Materials and Methods
2.1. Preparation of Composite Carbon Sources
2.2. Batch Experiment
2.3. Sampling and Analytical Methods
2.4. DNA Extraction, Quantitative Real-Time PCR (qPCR), and Illumina MiSeq Sequencing Analysis
2.5. Microbial Network Analysis
2.6. Statistical Analysis
3. Results and Discussion
3.1. Denitrification Performance, DOC Release, and N2O Emission of Different Composite Carbon Sources
3.2. Nitrogen Removal Performance, DOC Release, and N2O Accumulation Response to Variations of Different Factors
3.3. Characterization of PC-OH before and after Use
3.4. Nitrogen Functional Gene Analysis in Different SPD Systems
3.5. Microbial Community Structure
3.5.1. Bacterial and Fungal Community Structure
3.5.2. Co-Occurrence Network Analysis for Microbial Communities and Environmental Factors
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, H.; Paerl, H.W.; Qin, B.; Zhu, G.; Hall, N.S.; Wu, Y. Determining critical nutrient thresholds needed to control harmful cyanobacterial blooms in eutrophic Lake Taihu, China. Environ. Sci. Technol. 2015, 49, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Schipper, L.A.; Robertson, W.D.; Gold, A.J.; Jaynes, D.B.; Cameron, S.C. Denitrifying bioreactors-An approach for reducing nitrate loads to receiving waters. Ecol. Eng. 2010, 36, 1532–1543. [Google Scholar] [CrossRef]
- Wang, J.; Chu, L. Biological nitrate removal from water and wastewater by solid-phase denitrification process. Biotechnol. Adv. 2016, 34, 1103–1112. [Google Scholar] [CrossRef]
- Ashok, V.; Hait, S. Remediation of nitrate-contaminated water by solid-phase denitrification process—A review. Environ. Sci. Pollut. Res. 2015, 22, 8075–8093. [Google Scholar] [CrossRef]
- Feng, L.; Chen, K.; Han, D.; Zhao, J.; Lu, Y.; Yang, G.; Mu, J.; Zhao, X. Comparison of nitrogen removal and microbial properties in solid-phase denitrification systems for water purification with various pretreated lignocellulosic carriers. Bioresour. Technol. 2017, 224, 236–245. [Google Scholar] [CrossRef]
- Chu, L.; Wang, J. Denitrification of groundwater using PHBV blends in packed bed reactors and the microbial diversity. Chemosphere 2016, 155, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Tokiwa, Y.; Calabia, B.P.; Ugwu, C.U.; Aiba, S. Biodegradability of plastics. Int. J. Mol. Sci. 2009, 10, 3722–3742. [Google Scholar] [CrossRef]
- Yang, Z.; Sun, H.; Zhou, Q.; Zhao, L.; Wu, W. Nitrogen removal performance in pilot-scale solid-phase denitrification systems using novel biodegradable blends for treatment of waste water treatment plants effluent. Bioresour. Technol. 2020, 305, 122994. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.H.; Li, X.F.; Lin, J.H. Preparation and properties of biodegradable bamboo powder/polycaprolactone composites. J. For. Res. 2009, 20, 271–274. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, Y.Y.; Kim, T.H. A review on alkaline pretreatment technology for bioconversion of lignocellulosic biomass. Bioresour. Technol. 2016, 199, 42–48. [Google Scholar] [CrossRef]
- Jönsson, L.J.; Martín, C. Pretreatment of lignocellulose: Formation of inhibitory by-products and strategies for minimizing their effects. Bioresour. Technol. 2016, 199, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Zheng, X.; Xin, J.; Sun, Z.; Zheng, T. Selective enhancement and verification of woody biomass digestibility as a denitrification carbon source. Bioresour. Technol. 2017, 244, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Mosier, N.; Wyman, C.; Dale, B.; Elander, R.; Lee, Y.Y.; Holtzapple, M.; Ladisch, M. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour. Technol. 2005, 96, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Andlar, M.; Rezić, T.; Marđetko, N.; Kracher, D.; Ludwig, R.; Šantek, B. Lignocellulose degradation: An overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng. Life Sci. 2018, 18, 768–778. [Google Scholar] [CrossRef] [PubMed]
- APHA. Standard Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association/American Water Works Association/Water Environment Federation: Washington, DC, USA, 2005. [Google Scholar]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Taherzadeh, M.J.; Ruan, Y.; Deng, Y.; Chen, J.S.; Lu, H.F.; Xu, X.Y. Denitrification performance and microbial communities of solid-phase denitrifying reactors using poly (butylene succinate)/bamboo powder composite. Bioresour. Technol. 2020, 305, 123033. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, Y.; Lv, F.; Wang, H.; Tu, S. Biological denitrification in simulated groundwater using polybutylene succinate or polylactic acid-based composites as carbon source. Desalination Water Treat. 2016, 57, 9925–9932. [Google Scholar] [CrossRef]
- Ghafari, S.; Hasan, M.; Aroua, M.K. Bio-electrochemical removal of nitrate from water and wastewater-a review. Bioresour. Technol. 2008, 10, 3965–3974. [Google Scholar] [CrossRef]
- Yang, Z.; Sun, H.; Wu, W. Intensified simultaneous nitrification and denitrification performance in integrated packed bed bioreactors using PHBV with different dosing methods. Environ. Sci. Pollut. Res. 2020, 27, 21560–21569. [Google Scholar] [CrossRef]
- He, Q.; Feng, C.; Peng, T.; Chen, N.; Hu, Q.; Hao, C. Denitrification of synthetic nitrate-contaminated groundwater combined with rice washing drainage treatment. Ecol. Eng. 2016, 95, 152–159. [Google Scholar] [CrossRef]
- Sun, S.; Sun, S.; Cao, X.; Sun, R. The role of pretreatment in improving the enzymatic hydrolysis of lignocellulosic materials. Bioresour. Technol. 2016, 199, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Van Rijn, J.; Tal, Y.; Schreier, H.J. Denitrification in recirculating systems: Theory and applications. Aquac. Eng. 2006, 34, 364–376. [Google Scholar] [CrossRef]
- Kishida, N.; Kim, J.H.; Kimochi, Y.; Nishimura, O.; Sasaki, H.; Sudo, R. Effect of C/N ratio on nitrous oxide emission from swine wastewater treatment process. Water Sci. Technol. 2004, 49, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Adouani, N.; Lendormi, T.; Limousy, L.; Sire, O. Effect of the carbon source on N2O emissions during biological denitrification. Resour. Conserv. Recycl. 2010, 54, 299–302. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Choi, H.; Cho, K.S. Effects of carbon source, C/N ratio, nitrate, temperature, and pH on N2O emission and functional denitrifying genes during heterotrophic denitrification. J. Environ. Sci. Health Part A 2019, 54, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Greenan, C.M.; Moorman, T.B.; Parkin, T.B.; Kaspar, T.C.; Jaynes, D.B. Denitrification in wood chip bioreactors at different water flows. J. Environ. Qual. 2009, 38, 1664–1671. [Google Scholar] [CrossRef]
- Moorman, T.B.; Parkin, T.B.; Kaspar, T.C.; Jaynes, D.B. Denitrification activity, wood loss, and N2O emissions over 9 years from a wood chip bioreactor. Ecol. Eng. 2010, 36, 1567–1574. [Google Scholar] [CrossRef]
- Shen, Z.; Hu, J.; Wang, J.; Zhou, Y. Biological denitrification using starch/polycaprolactone blends as carbon source and biofilm support. Desalination Water Treat. 2015, 54, 609–615. [Google Scholar] [CrossRef]
- Hu, R.; Zheng, X.; Zheng, T.; Xin, J.; Wang, H.; Sun, Q. Effects of carbon availability in a woody carbon source on its nitrate removal behavior in solid-phase denitrification. J. Environ. Manag. 2019, 246, 832–839. [Google Scholar] [CrossRef]
- Cai, Y.; Zheng, Z.; Schäfer, F.; Stinner, W.; Yuan, X.; Wang, H.; Cui, Z.; Wang, X. A review about pretreatment of lignocellulosic biomass in anaerobic digestion: Achievement and challenge in Germany and China. J. Clean. Prod. 2021, 299, 126885. [Google Scholar] [CrossRef]
- Poh, L.S.; Jiang, X.; Zhang, Z.; Liu, Y.; Ng, W.J.; Zhou, Y. N2O accumulation from denitrification under different temperatures. Appl. Microbiol. Biotechnol. 2015, 99, 9215–9226. [Google Scholar] [CrossRef]
- Zhu, S.M.; Deng, Y.L.; Ruan, Y.J.; Guo, X.S.; Shi, M.M.; Shen, J.Z. Biological denitrification using poly(butylene succinate) as carbon source and biofilm carrier for recirculating aquaculture system effluent treatment. Bioresour. Technol. 2015, 192, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Lv, J.; Feng, J. Spectral characterization of four kinds of biodegradable plastics: Poly (lactic acid), poly (butylenes adipate-co-terephthalate), poly (hydroxybutyrate-co-hydroxyvalerate) and poly (butylenes succinate) with FTIR and raman spectroscopy. J. Polym. Environ. 2013, 21, 108–114. [Google Scholar] [CrossRef]
- Casas, A.; Alonso, M.V.; Oliet, M.; Rojo, E.; Rodriguez, F. FTIR analysis of lignin regenerated from Pinus radiata and Eucalyptus globulus woods dissolved in imidazolium-based ionic liquids. J. Chem. Technol. Biotechnol. 2012, 87, 472–480. [Google Scholar] [CrossRef]
- Ishola, M.M.; Millati, R.; Syamsiah, S.; Cahyanto, M.N.; Niklasson, C.; Taherzadeh, M.J. Structural changes of oil palm empty fruit bunch (OPEFB) after fungal and phosphoric acid pretreatment. Molecules 2012, 17, 14995–15012. [Google Scholar]
- Zhang, Y.; Ji, G.; Wang, R. Functional gene groups controlling nitrogen transformation rates in a groundwater-restoring denitrification biofilter under hydraulic retention time constraints. Ecol. Eng. 2016, 87, 45–52. [Google Scholar] [CrossRef]
- Cao, Y.; Green, P.G.; Holden, P.A. Microbial community composition and denitrifying enzyme activities in salt marsh sediments. Appl. Environ. Microbiol. 2008, 74, 7585–7595. [Google Scholar] [CrossRef]
- Rotthauwe, J.H.; Witzel, K.P.; Liesack, W. The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 1997, 63, 4704–4712. [Google Scholar] [CrossRef]
- Sinthusith, N.; Terada, A.; Hahn, M.; Noophan, P.; Munakata-Marr, J.; Figueroa, L.A. Identification and quantification of bacteria and archaea responsible for ammonia oxidation in different activated sludge of full-scale wastewater treatment plants. J. Environ. Sci. Health Part A 2015, 50, 169–175. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Li, Y.; Wu, J. Enhanced nitrogen removal and quantitative analysis of removal mechanism in multistage surface flow constructed wetlands for the large-scale treatment of swine wastewater. J. Environ. Manag. 2019, 246, 575–582. [Google Scholar] [CrossRef]
- Song, K.; Kang, H.; Zhang, L.; Mitsch, W.J. Seasonal and spatial variations of denitrification and denitrifying bacterial community structure in created riverine wetlands. Ecol. Eng. 2012, 38, 130–134. [Google Scholar] [CrossRef]
- Domeignoz-Horta, L.A.; Spor, A.; Bru, D.; Breuil, M.C.; Bizouard, F.; Léonard, J.; Philippot, L. The diversity of the N2O reducers matters for the N2O: N2 denitrification end-product ratio across an annual and a perennial cropping system. Front. Microbiol. 2015, 6, 971. [Google Scholar] [CrossRef]
- Zhao, S.; Zhou, J.; Yuan, D.; Wang, W.; Zhou, L.; Pi, Y.; Zhu, G. NirS-type N2O-producers and nosZ II-type N2O-reducers determine the N2O emission potential in farmland rhizosphere soils. J. Soils Sediments 2020, 20, 461–471. [Google Scholar] [CrossRef]
- Kong, X.; Duan, Y.; Schramm, A.; Eriksen, J.; Holmstrup, M.; Larsen, T.; Bol, R.; Petersen, S.O. Mitigating N2O emissions from clover residues by 3,4-dimethylpyrazole phosphate (DMPP) without adverse effects on the earthworm Lumbricus terrestris. Soil Biol. Biochem. 2017, 104, 95–107. [Google Scholar] [CrossRef]
- Saarenheimo, J.; Rissanen, A.J.; Arvola, L.; Nykänen, H.; Lehmann, M.F.; Tiirola, M. Genetic and environmental controls on nitrous oxide accumulation in lakes. PLoS ONE 2015, 10, e0121201. [Google Scholar] [CrossRef] [PubMed]
- Nakasaki, K.; Idemoto, Y.; Abe, M.; Rollon, A.P. Comparison of organic matter degradation and microbial community during thermophilic composting of two different types of anaerobic sludge. Bioresour. Technol. 2009, 100, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Mergaert, J.; Boley, A.; Cnockaert, M.C.; Müller, W.R.; Swings, J. Identity and potential functions of heterotrophic bacterial isolates from a continuous-upflow fixed-bed reactor for denitrification of drinking water with bacterial polyester as source of carbon and electron donor. Syst. Appl. Microbiol. 2001, 24, 303–310. [Google Scholar] [CrossRef]
- Zhuang, J.L.; Sun, X.; Zhao, W.Q.; Zhang, X.; Zhou, J.J.; Ni, B.J.; Liu, Y.D.; Shapleigh, J.P.; Li, W. The anammox coupled partial-denitrification process in an integrated granular sludge and fixed-biofilm reactor developed for mainstream wastewater treatment: Performance and community structure. Water Res. 2022, 210, 117964. [Google Scholar] [CrossRef]
- Zielińska, M.; Rusanowska, P.; Jarząbek, J.; Nielsen, J.L. Community dynamics of denitrifying bacteria in full-scale wastewater treatment plants. Environ. Technol. 2016, 37, 2358–2367. [Google Scholar] [CrossRef]
- Da Silva, M.L.B.; Cantão, M.E.; Mezzari, M.P.; Ma, J.; Nossa, C.W. Assessment of bacterial and archaeal community structure in swine wastewater treatment processes. Microb. Ecol. 2015, 70, 77–87. [Google Scholar] [CrossRef]
- Lee, S.H.; Li, C.; Heber, A.J.; Ni, J.; Huang, H. Biofiltration of a mixture of ethylene, ammonia, n-butanol, and acetone gases. Bioresour. Technol. 2013, 127, 366–377. [Google Scholar] [CrossRef] [PubMed]
- MacRae, J.D.; Smit, J.O.H.N. Characterization of caulobacters isolated from wastewater treatment systems. Appl. Environ. Microbiol. 1991, 57, 751–758. [Google Scholar] [CrossRef]
- Wei, W.; Isobe, K.; Shiratori, Y.; Nishizawa, T.; Ohte, N.; Otsuka, S.; Senoo, K. N2O emission from cropland field soil through fungal denitrification after surface applications of organic fertilizer. Soil Biol. Biochem. 2014, 69, 157–167. [Google Scholar] [CrossRef]
- Castillo, J.M.; Nogales, R.; Romero, E. Biodegradation of 3,4 dichloroaniline by fungal isolated from the preconditioning phase of winery wastes subjected to vermicomposting. J. Hazard. Mater. 2014, 267, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Soni, S.K.; Soni, R. Regulation of cellulase synthesis in Chaetomium erraticum. BioResources 2010, 5, 81–98. [Google Scholar]
- Huang, Y.; Busk, P.K.; Lange, L. Cellulose and hemicellulose-degrading enzymes in Fusarium commune transcriptome and functional characterization of three identified xylanases. Enzyme Microb. Technol. 2015, 73, 9–19. [Google Scholar] [CrossRef]
- Novy, V.; Nielsen, F.; Seiboth, B.; Nidetzky, B. The influence of feedstock characteristics on enzyme production in Trichoderma reesei: A review on productivity, gene regulation and secretion profiles. Biotechnol. Biofuels 2019, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Malafatti-Picca, L.; de Barros Chaves, M.R.; de Castro, A.M.; Valoni, É.; de Oliveira, V.M.; Marsaioli, A.J.; de Franceschi de Angelis, D.; Attili-Angelis, D. Hydrocarbon-associated substrates reveal promising fungi for poly (ethylene terephthalate) (PET) depolymerization. Braz. J. Microbiol. 2019, 50, 633–648. [Google Scholar] [CrossRef]
- Gao, P.; Gao, Y.; Wang, H.; Ma, T.; Gu, J.D. An evaluation of different detection methods for anaerobic ammonium-oxidizing (anammox) bacteria inhabiting the oil reservoir systems. Int. Biodeterior. Biodegradation 2023, 177, 105536. [Google Scholar] [CrossRef]
- Li, S.; Wu, F. Diversity and co-occurrence patterns of soil bacterial and fungal communities in seven intercropping systems. Front. Microbiol. 2018, 9, 1521. [Google Scholar] [CrossRef]
- Shi, Y.; Delgado-Baquerizo, M.; Li, Y.; Yang, Y.; Zhu, Y.G.; Peñuelas, J.; Chu, H. Abundance of kinless hubs within soil microbial networks are associated with high functional potential in agricultural ecosystems. Environ. Int. 2020, 142, 105869. [Google Scholar] [CrossRef] [PubMed]
- Francis, C.A.; Roberts, K.J.; Beman, J.M.; Santoro, A.E.; Oakley, B.B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 2005, 102, 14683–14688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsushima, I.; Kindaichi, T.; Okabe, S. Quantification of anaerobic ammonium-oxidizing bacteria in enrichment cultures by real-time PCR. Water Res. 2007, 41, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J. Habitat segregation of a functional gene encoding nitrate ammonification in estuarine sediments. Geomicrobiol. J. 2006, 23, 75–87. [Google Scholar] [CrossRef]
- López-Gutiérrez, J.C.; Henry, S.; Hallet, S.; Martin-Laurent, F.; Catroux, G.; Philippot, L. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 2004, 57, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Bru, D.; Sarr, A.; Philippot, L. Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl. Environ. Microbiol. 2007, 73, 5971–5974. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tiquia, S.M.; Holguin, G.; Wu, L.; Nold, S.C.; Devol, A.H.; Luo, K.; Palumbo, A.V.; Tiedje, J.M.; Zhou, J. Molecular diversity of denitrifying genes in continental margin sediments within the oxygen-deficient zone off the Pacific coast of Mexico. Appl. Environ. Microbiol. 2003, 69, 3549–3560. [Google Scholar] [CrossRef]
- Kandeler, E.; Deiglmayr, K.; Tscherko, D.; Bru, D.; Philippot, L. Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl. Environ. Microbiol. 2006, 72, 5957–5962. [Google Scholar] [CrossRef]
- Li, X.; Wang, H.; Hu, C.; Yang, M.; Hu, H.; Niu, J. Characteristics of biofilms and iron corrosion scales with ground and surface waters in drinking water distribution systems. Corros Sci 2015, 90, 331–339. [Google Scholar] [CrossRef]
- Jones, C.M.; Graf, D.R.; Bru, D.; Philippot, L.; Hallin, S. The unaccounted yet abundant nitrous oxide-reducing microbial community: A potential nitrous oxide sink. ISME J. 2013, 7, 417–426. [Google Scholar] [CrossRef]
- Muyzer, G.; De Waal, E.C.; Uitterlinden, A. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, S.; Jiang, Y.; Li, M.; Yuan, J.; Wang, G. Influence mechanism of filling ratio on solid-phase denitrification with polycaprolactone as biofilm carrier. Bioresour. Technol. 2021, 337, 125401. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, G.; Wang, Y.; Wang, S.; Yin, C. Nitrous oxide reductase gene (nosZ) and N2O reduction along the littoral gradient of a eutrophic freshwater lake. J. Environ. Sci. 2013, 25, 44–52. [Google Scholar] [CrossRef]
- Zhou, B.; Duan, J.; Xue, L.; Zhang, J.; Yang, L. Effect of plant-based carbon source supplements on denitrification of synthetic wastewater: Focus on the microbiology. Environ. Sci. Pollut. Res. 2019, 26, 24683–24694. [Google Scholar] [CrossRef]
- Li, H.; Zhou, Z.; Liu, Q.; Dong, H.; Duan, Y.; Li, C.; Zhang, J.; Tan, H. Biological denitrification in high salinity wastewater using semen litchi as a carbon source. Rsc Adv. 2015, 5, 92836–92842. [Google Scholar] [CrossRef]
- Kundu, P.; Pramanik, A.; Dasgupta, A.; Mukherjee, S.; Mukherjee, J. Simultaneous heterotrophic nitrification and aerobic denitrification by Chryseobacterium sp. R31 isolated from abattoir wastewater. BioMed Res. Int. 2014, 2014, 436056. [Google Scholar] [CrossRef] [PubMed]
- Greening, C.; Geier, R.; Wang, C.; Woods, L.C.; Morales, S.E.; McDonald, M.J.; Rushton-Green, R.; Morgan, X.C.; Koike, S.; Leahy, S.C.; et al. Alternative hydrogen uptake pathways suppress methane production in ruminants. bioRxiv 2018. [Google Scholar] [CrossRef]
- Xiao, Y.; Zheng, Y.; Wu, S.; Yang, Z.H.; Zhao, F. Bacterial community structure of autotrophic denitrification biocathode by 454 pyrosequencing of the 16S rRNA gene. Microb. Ecol. 2015, 69, 492–499. [Google Scholar] [CrossRef]
- Miller, M.N.; Dandie, C.E.; Zebarth, B.J.; Burton, D.L.; Goyer, C.; Trevors, J.T. Influence of carbon amendments on soil denitrifier abundance in soil microcosms. Geoderma 2012, 170, 48–55. [Google Scholar] [CrossRef]
- Wang, H.; Sun, Y.; Zhang, L.; Wang, W.; Guan, Y. Enhanced nitrogen removal and mitigation of nitrous oxide emission potential in a lab-scale rain garden with internal water storage. J. Water Process. Eng. 2021, 42, 102147. [Google Scholar] [CrossRef]
- Xie, C.H.; Yokota, A. Reclassification of [Flavobacterium] ferrugineum as Terrimonas ferruginea gen. nov., comb. nov., and description of Terrimonas lutea sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2006, 56, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Hetz, S.A.; Horn, M.A. Burkholderiaceae are key acetate assimilators during complete denitrification in acidic cryoturbated peat circles of the arctic tundra. Front. Microbiol. 2021, 12, 628269. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, J.; Li, C.; Deng, Y.; Zhang, Z.; Ye, Z.; Zhu, S. Poly (butylene succinate)/bamboo powder blends as solid-phase carbon source and biofilm carrier for denitrifying biofilters treating wastewater from recirculating aquaculture system. Sci. Rep. 2018, 8, 3289. [Google Scholar] [CrossRef]
- Bonato, P.; Batista, M.B.; Camilios-Neto, D.; Pankievicz, V.C.; Tadra-Sfeir, M.Z.; Monteiro, R.A.; Pedrosa, F.O.; Souza, E.M.; Chubatsu, L.S.; Wassem, R.; et al. RNA-seq analyses reveal insights into the function of respiratory nitrate reductase of the diazotroph Herbaspirillum seropedicae. Environ. Microbiol. 2016, 18, 2677–2688. [Google Scholar] [CrossRef]
- King, G.M. Nitrate-dependent anaerobic carbon monoxide oxidation by aerobic CO-oxidizing bacteria. FEMS Microbiol. Ecol. 2006, 56, 1–7. [Google Scholar] [CrossRef]
- Qiu, Y.L.; Kuang, X.Z.; Shi, X.S.; Yuan, X.Z.; Guo, R.B. Paludibacter jiangxiensis sp. nov., a strictly anaerobic, propionate-producing bacterium isolated from rice paddy field. Arch. Microbiol. 2014, 196, 149–155. [Google Scholar] [CrossRef]
- Soares, L.A.; Silva Rabelo, C.A.B.; Sakamoto, I.K.; Silva, E.L.; Varesche, M.B.A. Screening and bioprospecting of anaerobic consortia for biofuel production enhancement from sugarcane bagasse. Appl. Biochem. Biotechnol. 2020, 190, 232–251. [Google Scholar] [CrossRef] [PubMed]
- Song, W.F.; Wang, J.W.; Yan, Y.C.; An, L.Y.; Zhang, F.; Wang, L.; Xu, Y.; Tian, M.Z.; Nie, Y.; Wu, X.L. Shifts of the indigenous microbial communities from reservoir production water in crude oil-and asphaltene-degrading microcosms. Int. Biodeterior. Biodegrad. 2018, 132, 18–29. [Google Scholar] [CrossRef]
- Rusznyak, A.; Toth, E.M.; Schumann, P.; Spröer, C.; Makk, J.; Szabo, G.; Vladar, P.; Marialigeti, K.; Borsodi, A.K. Cellulomonas phragmiteti sp. nov., a cellulolytic bacterium isolated from reed (Phragmites australis) periphyton in a shallow soda pond. Int. J. Syst. Evol. Microbiol. 2011, 61, 1662–1666. [Google Scholar] [CrossRef]
- Xafenias, N.; Anunobi, M.O.; Mapelli, V. Electrochemical startup increases 1, 3-propanediol titers in mixed-culture glycerol fermentations. Process Biochem. 2015, 50, 1499–1508. [Google Scholar] [CrossRef]
- Chang, J.J.; Lin, J.J.; Ho, C.Y.; Chin, W.C.; Huang, C.C. Establishment of rumen-mimic bacterial consortia: A functional union for bio-hydrogen production from cellulosic bioresource. Int. J. Hydrogen Energy 2010, 35, 13399–13406. [Google Scholar] [CrossRef]
- Wang, J.; Wang, C.; Li, J.; Bai, P.; Li, Q.; Shen, M.; Li, R.; Li, T.; Zhao, J. Comparative genomics of degradative Novosphingobium strains with special reference to microcystin-degrading Novosphingobium sp. THN1. Front. Microbiol. 2018, 9, 2238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- S. Steindorff, A.S.; Serra, L.A.; Formighieri, E.F.; de Faria, F.P.; Poças-Fonseca, M.J.; de Almeida, J.R.M. Insights into the Lignocellulose-Degrading Enzyme System of Humicola grisea var. thermoidea Based on Genome and Transcriptome Analysis. Microbiol. Spectr. 2021, 9, e01088-21. [Google Scholar] [CrossRef] [PubMed]
- Štursová, M.; Žifčáková, L.; Leigh, M.B.; Burgess, R.; Baldrian, P. Cellulose utilization in forest litter and soil: Identification of bacterial and fungal decomposers. FEMS Microbiol. Ecol. 2012, 80, 735–746. [Google Scholar] [CrossRef]
- Ali, A.; Ellinger, B.; Brandt, S.C.; Betzel, C.; Rühl, M.; Wrenger, C.; Schlüter, H.; Schäfer, W.; Brognaro, H.; Gand, M. Genome and Secretome Analysis of Staphylotrichum longicolleum DSM105789 Cultured on Agro-Residual and Chitinous Biomass. Microorganisms 2021, 9, 1581. [Google Scholar] [CrossRef]
- Tian, X.; Yang, T.; He, J.; Chu, Q.; Jia, X.; Huang, J. Fungal community and cellulose-degrading genes in the composting process of Chinese medicinal herbal residues. Bioresour. Technol. 2017, 241, 374–383. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Lou, Y.; Pan, H.; Wang, H.; Yang, Q.; Zhuge, Y.; Hu, J. Improved Denitrification Performance of Polybutylene Succinate/Corncob Composite Carbon Source by Proper Pretreatment: Performance, Functional Genes and Microbial Community Structure. Polymers 2023, 15, 801. https://doi.org/10.3390/polym15040801
Yang Z, Lou Y, Pan H, Wang H, Yang Q, Zhuge Y, Hu J. Improved Denitrification Performance of Polybutylene Succinate/Corncob Composite Carbon Source by Proper Pretreatment: Performance, Functional Genes and Microbial Community Structure. Polymers. 2023; 15(4):801. https://doi.org/10.3390/polym15040801
Chicago/Turabian StyleYang, Zhongchen, Yanhong Lou, Hong Pan, Hui Wang, Quangang Yang, Yuping Zhuge, and Jingying Hu. 2023. "Improved Denitrification Performance of Polybutylene Succinate/Corncob Composite Carbon Source by Proper Pretreatment: Performance, Functional Genes and Microbial Community Structure" Polymers 15, no. 4: 801. https://doi.org/10.3390/polym15040801