Calcium-Alginate-Chitosan Nanoparticle as a Potential Solution for Pesticide Removal, a Computational Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pesticide Database Preparation
- We only considered compounds that were registered with the Chemical Abstracts Service (CAS).
- To conduct a comprehensive search, we utilized the PubChem database [57] and selected the advanced search option to filter compounds by their unique CAS identifier.
- Using the RDKit python library (RDKit: Open-source cheminformatics. https://www.rdkit.org, accessed on 3 March 2023), we processed all compounds in the final database by generating 100 conformers per molecule. Each conformer was evaluated using the ETKDGv3 conformer generator [58], a stochastic search method that combines distance geometry with knowledge obtained from experimental crystal structures.
- To maintain uniformity in our compound preparations, we employed OpenBabel software [59] to adjust the pH of all samples to a range of 7.0 ± 2.0.
2.2. Calcium-Alginate-Chitosan Nanoparticle Preparation
2.3. Ensemble Conformers Selection
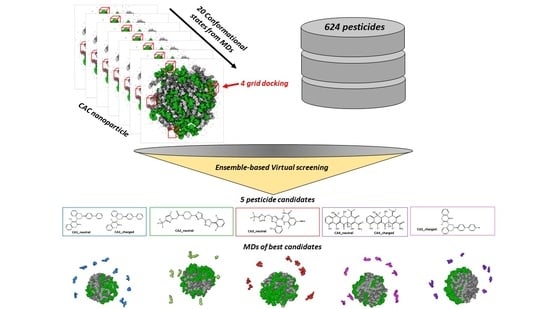
2.4. Ensemble-Based Virtual Screening
2.5. Pesticides Capture Studies
2.6. Non-Covalent Interactions
2.7. Adsorbent and Absorbent Characterization
3. Results and Discussions
3.1. Pesticide Database Characterization
3.2. Nanoparticle Formation
3.3. Virtual Screening Analysis
3.4. Capture Analysis
3.5. Non-Covalent Interactions Analysis
3.6. Adsorption and Absorption Characterization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rani, L.; Thapa, K.; Kanojia, N.; Sharma, N.; Singh, S.; Grewal, A.S.; Srivastav, A.L.; Kaushal, J. An extensive review on the consequences of chemical pesticides on human health and environment. J. Clean. Prod. 2021, 283, 124657. [Google Scholar] [CrossRef]
- Kim, K.-H.; Kabir, E.; Jahan, S.A. Exposure to pesticides and the associated human health effects. Sci. Total Environ. 2017, 575, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Alengebawy, A.; Abdelkhalek, S.T.; Qureshi, S.R.; Wang, M.-Q. Heavy Metals and Pesticides Toxicity in Agricultural Soil and Plants: Ecological Risks and Human Health Implications. Toxics 2021, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Sarwar, M. The Dangers of Pesticides Associated with Public Health and Preventing of the Risks. Int. J. Bioinform. Biomed. Eng. 2015, 1, 130–136. [Google Scholar]
- Sánchez-Bayo, F. Indirect effect of pesticides on insects and other arthropods. Toxics 2021, 9, 177. [Google Scholar] [CrossRef]
- Walker, C.H. Neurotoxic pesticides and behavioural effects upon birds. Ecotoxicology 2003, 12, 307–316. [Google Scholar] [CrossRef]
- Fry, D.M. Reproductive effects in birds exposed to pesticides and industrial chemicals. Environ. Health Perspect. 1995, 103 (Suppl. 7), 165–171. [Google Scholar] [CrossRef]
- Paoli, D.; Giannandrea, F.; Gallo, M.; Turci, R.; Cattaruzza, M.S.; Lombardo, F.; Lenzi, A.; Gandini, L. Exposure to polychlorinated biphenyls and hexachlorobenzene, semen quality and testicular cancer risk. J. Endocrinol. Investig. 2015, 38, 745–752. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, F.P. Pesticides, environment, and food safety. Food Energy Secur. 2017, 6, 48–60. [Google Scholar] [CrossRef]
- Van Maele-Fabry, G.; Lantin, A.-C.; Hoet, P.; Lison, D. Childhood leukaemia and parental occupational exposure to pesticides: A systematic review and meta-analysis. Cancer Causes Control. 2010, 21, 787–809. [Google Scholar] [CrossRef]
- Mostafalou, S.; Abdollahi, M. Pesticides and human chronic diseases: Evidences, mechanisms, and perspectives. Toxicol. Appl. Pharmacol. 2013, 268, 157–177. [Google Scholar] [CrossRef]
- Richardson, J.R.; Fitsanakis, V.; Westerink, R.H.S.; Kanthasamy, A.G. Neurotoxicity of pesticides. Acta Neuropathol. 2019, 138, 343–362. [Google Scholar] [CrossRef]
- El-Nahhal, Y.; El-Nahhal, I. Cardiotoxicity of some pesticides and their amelioration. Environ. Sci. Pollut. Res. 2021, 28, 44726–44754. [Google Scholar] [CrossRef]
- Boedeker, W.; Watts, M.; Clausing, P.; Marquez, E. The global distribution of acute unintentional pesticide poisoning: Estimations based on a systematic review. BMC Public Health 2020, 20, 1875. [Google Scholar] [CrossRef]
- Zohry, A.E.-H.; Ouda, S. Crop Rotation Defeats Pests and Weeds; No. Hald 1999; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar] [CrossRef]
- Ndakidemi, B.J.; Mbega, E.R.; Ndakidemi, P.A.; Stevenson, P.C.; Belmain, S.R.; Arnold, S.E.J.; Woolley, V.C. Natural pest regulation and its compatibility with other crop protection practices in smallholder bean farming systems. Biology 2021, 10, 805. [Google Scholar] [CrossRef]
- Kumari, P.; Jasrotia, P.; Kumar, D.; Kashyap, P.L.; Kumar, S.; Mishra, C.N.; Kumar, S.; Singh, G.P. Biotechnological Approaches for Host Plant Resistance to Insect Pests. Front. Genet. 2022, 13, 914029. [Google Scholar] [CrossRef]
- Andrunik, M.; Bajda, T. Removal of pesticides from waters by adsorption: Comparison between synthetic zeolites and mesoporous silica materials. A review. Materials 2021, 14, 3532. [Google Scholar] [CrossRef]
- Almakhathi, A.A.S.; Zeeshan, M.; Shah, J.; Jan, M.R. Simultaneous Removal and Extraction of Bisphenol A and 4-tert-butylphenol From Water Samples Using Magnetic Chitosan Particles. Front. Mater. 2022, 9, 786581. [Google Scholar] [CrossRef]
- Sanoja-López, K.A.; Quiroz-Suárez, K.A.; Dueñas-Rivadeneira, A.A.; Maddela, N.R.; Montenegro, M.C.M.; Luque, R.; Rodríguez-Díaz, J.M. Polymeric membranes functionalized with nanomaterials (MP@NMs): A review of advances in pesticide removal. Environ. Res. 2023, 217, 114776. [Google Scholar] [CrossRef]
- Kyriakopoulos, G.; Doulia, D. Adsorption of pesticides on carbonaceous and polymeric materials from aqueous solutions: A review. Sep. Purif. Rev. 2006, 35, 97–191. [Google Scholar] [CrossRef]
- Morales-Quintana, L.; Bustos, D.; Gallego, J.; Valdes, O.; Guzmán, L.; Nachtigall, F.M.; Marican, A.; Hernández-Rodríguez, E.W.; Avila-Salas, F.; Bueno-Silva, W.; et al. Physicochemical and computational analysis of the melamine resin derivative for the glyphosate absorption from water using Langmuir-type model. Int. J. Environ. Sci. Technol. 2022, 19, 7791–7802. [Google Scholar] [CrossRef]
- Escudero, L.B.; Quintas, P.Y.; Wuilloud, R.G.; Dotto, G.L. Recent advances on elemental biosorption. Environ. Chem. Lett. 2019, 17, 409–427. [Google Scholar] [CrossRef]
- Berger, J.; Reist, M.; Mayer, J.; Felt, O.; Peppas, N.; Gurny, R. Structure and interactions in covalently and ionically crosslinked chitosan hydrogels for biomedical applications. Eur. J. Pharm. Biopharm. 2004, 57, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Bartkowiak, A.; Hunkeler, D. Alginate-Oligochitosan Microcapsules: A Mechanistic Study Relating Membrane and Capsule Properties to Reaction Conditions. Chem. Mater. 1999, 11, 2486–2492. [Google Scholar] [CrossRef]
- Kulig, D.; Zimoch-Korzycka, A.; Jarmoluk, A.; Marycz, K. Study on alginate-chitosan complex formed with different polymers ratio. Polymers 2016, 8, 167. [Google Scholar] [CrossRef]
- Jiménez-Gómez, C.P.; Cecilia, J.A. Chitosan: A Natural Biopolymer with a Wide and Varied Range of Applications. Molecules 2020, 25, 3981. [Google Scholar] [CrossRef]
- Bagheri-Khoulenjani, S.; Taghizadeh, S.; Mirzadeh, H. An investigation on the short-term biodegradability of chitosan with various molecular weights and degrees of deacetylation. Carbohydr. Polym. 2009, 78, 773–778. [Google Scholar] [CrossRef]
- Rodrigues, S.; Dionísio, M.; López, C.R.; Grenha, A. Biocompatibility of Chitosan Carriers with Application in Drug Delivery. J. Funct. Biomater. 2012, 3, 615–641. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.-Y.; Zhu, J.-F. Study on antimicrobial activity of chitosan with different molecular weights. Carbohydr. Polym. 2003, 54, 527–530. [Google Scholar] [CrossRef]
- Xia, W.; Liu, P.; Zhang, J.; Chen, J. Biological activities of chitosan and chitooligosaccharides. Food Hydrocoll. 2011, 25, 170–179. [Google Scholar] [CrossRef]
- Silva, A.O.; Cunha, R.S.; Hotza, D.; Machado, R.A.F. Chitosan as a matrix of nanocomposites: A review on nanostructures, processes, properties, and applications. Carbohydr. Polym. 2021, 272, 118472. [Google Scholar] [CrossRef]
- Sirajudheen, P.; Poovathumkuzhi, N.C.; Vigneshwaran, S.; Chelaveettil, B.M.; Meenakshi, S. Applications of chitin and chitosan based biomaterials for the adsorptive removal of textile dyes from water—A comprehensive review. Carbohydr. Polym. 2021, 273, 118604. [Google Scholar] [CrossRef]
- Khor, E.; Lim, L.Y. Implantable applications of chitin and chitosan. Biomaterials 2003, 24, 2339–2349. [Google Scholar] [CrossRef]
- Castro, R.I.; Morales-Quintana, L.; Alvarado, N.; Guzmán, L.; Forero-Doria, O.; Valenzuela-Riffo, F.; Laurie, V.F. Design and optimization of a self-assembling complex based on microencapsulated calcium alginate and glutathione (Cag) using response surface methodology. Polymers 2021, 13, 2080. [Google Scholar] [CrossRef]
- Makarova, A.O.; Derkach, S.R.; Khair, T.; Kazantseva, M.A.; Zuev, Y.F.; Zueva, O.S. Ion-Induced Polysaccharide Gelation: Peculiarities of Alginate Egg-Box Association with Different Divalent Cations. Polymers 2023, 15, 1243. [Google Scholar] [CrossRef]
- Hu, C.; Lu, W.; Mata, A.; Nishinari, K.; Fang, Y. Ions-induced gelation of alginate: Mechanisms and applications. Int. J. Biol. Macromol. 2021, 177, 578–588. [Google Scholar] [CrossRef]
- Hu, C.; Lu, W.; Sun, C.; Zhao, Y.; Zhang, Y.; Fang, Y. Gelation behavior and mechanism of alginate with calcium: Dependence on monovalent counterions. Carbohydr. Polym. 2022, 294, 119788. [Google Scholar] [CrossRef]
- Hecht, H.; Srebnik, S. Structural Characterization of Sodium Alginate and Calcium Alginate. Biomacromolecules 2016, 17, 2160–2167. [Google Scholar] [CrossRef]
- Bustos, D.; Hernández-Rodríguez, E.W.; Castro, R.I.; Morales-Quintana, L. Structural Effects of pH Variation and Calcium Amount on the Microencapsulation of Glutathione in Alginate Polymers. Biomed. Res. Int. 2022, 2022, 5576090. [Google Scholar] [CrossRef]
- Marwah, H.; Khare, S.; Rawat, P.; Singh, S.; Kesharwani, P.; Alam, M.S.; Hamid, H.; Arora, S. Strengths, limitations, and regulatory aspects of hybrid drug delivery systems. Hybrid Nanomater. Drug Deliv. 2022, 339–355. [Google Scholar] [CrossRef]
- Ahmad, M.F.; Haydar, S.; Bhatti, A.A.; Bari, A.J. Application of artificial neural network for the prediction of biosorption capacity of immobilized Bacillus subtilis for the removal of cadmium ions from aqueous solution. Biochem. Eng. J. 2014, 84, 83–90. [Google Scholar] [CrossRef]
- Zang, T.; Cheng, Z.; Lu, L.; Jin, Y.; Xu, X.; Ding, W.; Qu, J. Removal of Cr(VI) by modified and immobilized Auricularia auricula spent substrate in a fixed-bed column. Ecol. Eng. 2017, 99, 358–365. [Google Scholar] [CrossRef]
- Boddu, V.M.; Abburi, K.; Talbott, J.L.; Smith, E.D.; Haasch, R. Removal of arsenic (III) and arsenic (V) from aqueous medium using chitosan-coated biosorbent. Water Res. 2008, 42, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Cadaval, T.R.S., Jr.; Dotto, G.L.; Seus, E.R.; Mirlean, N.; de Almeida Pinto, L.A. Vanadium removal from aqueous solutions by adsorption onto chitosan films. Desalination Water Treat. 2015, 57, 16583–16591. [Google Scholar] [CrossRef]
- Liu, X.-Q.; Zhao, X.-X.; Liu, Y.; Zhang, T.-A. Review on preparation and adsorption properties of chitosan and chitosan composites. Polym. Bull. 2022, 79, 2633–2665. [Google Scholar] [CrossRef]
- Vieira, M.; Esquerdo, V.; Nobre, L.; Dotto, G.; Pinto, L. Glass beads coated with chitosan for the food azo dyes adsorption in a fixed bed column. J. Ind. Eng. Chem. 2014, 20, 3387–3393. [Google Scholar] [CrossRef]
- Alsamman, M.T.; Sánchez, J. Recent advances on hydrogels based on chitosan and alginate for the adsorption of dyes and metal ions from water. Arab. J. Chem. 2021, 14, 103455. [Google Scholar] [CrossRef]
- Semeraro, P.; Fini, P.; D’addabbo, M.; Rizzi, V.; Cosma, P. Removal from wastewater and recycling of azo textile dyes by alginate-chitosan beads. Int. J. Environ. Agric. Biotechnol. (IJEAB) 2017, 2, 238860. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, X.; Lou, T. Preparation of fibrous chitosan/sodium alginate composite foams for the adsorption of cationic and anionic dyes. J. Hazard. Mater. 2021, 403, 124054. [Google Scholar] [CrossRef]
- Valdés, C.; Valdés, O.; Bustos, D.; Abril, D.; Cabrera-Barjas, G.; Pereira, A.; Villaseñor, J.; Polo-Cuadrado, E.; Carreño, G.; Durán-Lara, E.F.; et al. Use of poly(Vinyl alcohol)-malic acid (CLHPMA) hydrogels and chitosan coated calcium alginate (CCCA) microparticles as potential sorbent phases for the extraction and quantitative determination of pesticides from aqueous solutions. Polymers 2021, 13, 3993. [Google Scholar] [CrossRef]
- Lu, L.C.; Wang, C.I.; Sye, W.F. Applications of chitosan beads and porous crab shell powder for the removal of 17 organochlorine pesticides (OCPs) in water solution. Carbohydr. Polym. 2011, 83, 1984–1989. [Google Scholar] [CrossRef]
- Etcheverry, M.; Cappa, V.; Trelles, J.; Zanini, G. Montmorillonite-alginate beads: Natural mineral and biopolymers based sorbent of paraquat herbicides. J. Environ. Chem. Eng. 2017, 5, 5868–5875. [Google Scholar] [CrossRef]
- El Harmoudi, H.; El Gaini, L.; Daoudi, E.; Rhazi, M.; Boughaleb, Y.; El Mhammedi, M.; Migalska-Zalas, A.; Bakasse, M. Removal of 2,4-D from aqueous solutions by adsorption processes using two biopolymers: Chitin and chitosan and their optical properties. Opt. Mater. 2014, 36, 1471–1477. [Google Scholar] [CrossRef]
- Cabrera-Barjas, G.; Gallardo, F.; Nesic, A.; Taboada, E.; Marican, A.; Mirabal-Gallardo, Y.; Avila-Salas, F.; Delgado, N.; de Armas-Ricard, M.; Valdes, O. Utilization of industrial by-product fungal biomass from Aspergillus niger and Fusarium culmorum to obtain biosorbents for removal of pesticide and metal ions from aqueous solutions. J. Environ. Chem. Eng. 2020, 8, 104355. [Google Scholar] [CrossRef]
- Nadavala, S.K.; Swayampakula, K.; Boddu, V.M.; Abburi, K. Biosorption of phenol and o-chlorophenol from aqueous solutions on to chitosan–calcium alginate blended beads. J. Hazard. Mater. 2009, 162, 482–489. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Wang, S.; Witek, J.; Landrum, G.A.; Riniker, S. Improving Conformer Generation for Small Rings and Macrocycles Based on Distance Geometry and Experimental Torsional-Angle Preferences. J. Chem. Inf. Model. 2020, 60, 2044–2058. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open chemical toolbox. J. Cheminformatics 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pKa prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Li, J.; Abel, R.; Zhu, K.; Cao, Y.; Zhao, S.; Friesner, R.A. The VSGB 2.0 Model: A Next Generation Energy Model for High Resolution Protein Structure Modeling. Proteins 2011, 79, 2794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maestro, S. Schrödinger Release 2021-1; LLC: New York, NY, USA, 2020. [Google Scholar]
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.J.; Skjærven, L.; Yao, X.Q. The Bio3D packages for structural bioinformatics. Protein Sci. 2021, 30, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Tang, S.; Mei, Z.; Wang, L.; Huang, Q.; Hu, H.; Ling, M.; Wu, J. Vina-GPU 2.0: Further Accelerating AutoDock Vina and Its Derivatives with Graphics Processing Units. J. Chem. Inf. Model. 2023, 63, 1982–1998. [Google Scholar] [CrossRef]
- Bajusz, D.; Rácz, A.; Héberger, K. Comparison of data fusion methods as consensus scores for ensemble docking. Molecules 2019, 24, 2690. [Google Scholar] [CrossRef] [Green Version]
- Boto, R.; Peccati, F.; Laplaza, R.; Quan, C.; Carbone, A.; Piquemal, J.-P.; Maday, Y.; Contreras-García, J. NCIPLOT4: Fast, Robust, and Quantitative Analysis of Noncovalent Interactions. J. Chem. Theory Comput. 2020, 16, 4150–4158. [Google Scholar] [CrossRef]
- Giorgino, T. Computing 1-D atomic densities in macromolecular simulations: The density profile tool for VMD. Comput. Phys. Commun. 2014, 185, 317–322. [Google Scholar] [CrossRef] [Green Version]
- Giorgino, T. Computing diffusion coefficients in macromolecular simulations: The Diffusion Coefficient Tool for VMD. J. Open Source Softw. 2019, 4, 1698. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Damin-Pernik, M.; Espana, B.; Besse, S.; Fourel, I.; Caruel, H.; Popowycz, F.; Benoit, E.; Lattard, V. Development of an Ecofriendly Anticoagulant Rodenticide Based on the Stereochemistry of Difenacoum. Drug Metab. Dispos. 2016, 44, 1872–1880. [Google Scholar] [CrossRef] [Green Version]
- Khalil, R.A.; Barbier, B.; Rached, A.; Benoit, E.; Pinot, A.; Lattard, V. Water vole management—Could anticoagulant rodenticides stereochemistry mitigate the ecotoxicity issues associated to their use? Environ. Toxicol. Pharmacol. 2021, 81, 103536. [Google Scholar] [CrossRef]
- Zhang, F.; Lv, X.; Jia, H.; Huang, C.; Wei, J.; Ding, Z.; Wang, F.; Wang, J. Toxicity of the novel fungicide oxathiapiprolin to Chlorella vulgaris: Assessments at different levels of biological organization. Chemosphere 2022, 291, 132752. [Google Scholar] [CrossRef]
- Zhou, L.; Wu, Q.; Gao, Y.; Shi, H.; Wang, M. Enantioselective aquatic toxicity and degradation in soil of the chiral fungicide oxathiapiprolin. Sci. Total Environ. 2022, 836, 155632. [Google Scholar] [CrossRef]
- Ma, D.; Yang, S.; Jiang, J.; Zhu, J.; Li, B.; Mu, W.; Dou, D.; Liu, F. Toxicity, residue and risk assessment of tetraniliprole in soil-earthworm microcosms. Ecotoxicol. Environ. Saf. 2021, 213, 112061. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, X.; Liu, Y.; Fang, K.; Liu, T. Residue and toxicity of cyantraniliprole and its main metabolite J9Z38 in soil-earthworm microcosms. Chemosphere 2020, 249, 126479. [Google Scholar] [CrossRef]
- Zhang, H.; Song, X.; Zhang, J.; Liu, Y.; Zhao, H.; Hu, J.; Zhao, J. Performance and mechanism of sycamore flock based biochar in removing oxytetracycline hydrochloride. Bioresour. Technol. 2022, 350, 126884. [Google Scholar] [CrossRef]
- Li, X.; Gan, T.; Zhang, J.; Shi, Z.; Liu, Z.; Xiao, Z. High-capacity removal of oxytetracycline hydrochloride from wastewater via Mikania micrantha Kunth-derived biochar modified by Zn/Fe-layered double hydroxide. Bioresour. Technol. 2022, 361, 127646. [Google Scholar] [CrossRef]
- Rached, A.; Lattard, V.; Fafournoux, A.; Caruel, H.; Fourel, I.; Benoit, E.; Lefebvre, S. Comparative pharmacokinetics of difethialone stereoisomers in male and female rats and mice: Development of an intra- and inter-species model to predict the suitable formulation mix. Arch. Toxicol. 2022, 96, 535–544. [Google Scholar] [CrossRef]
- Erdem, S.; Öztekin, M.; Açıkel, Y.S. Investigation of tetracycline removal from aqueous solutions using halloysite/chitosan nanocomposites and halloysite nanotubes/alginate hydrogel beads. Environ. Nanotechnol. Monit. Manag. 2021, 16, 100576. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, Y.; Li, L.; Jia, L. Fabrication of alginate-based nanofibers loaded with ZnO nanoparticles for adsorption of tetracyclines from environmental waters. Mater Today Commun. 2023, 34, 105214. [Google Scholar] [CrossRef]







| Rule | Formula |
|---|---|
| Minimum value | |
| Arithmetic Mean | |
| Geometric Mean | |
| Harmonic Mean | |
| Median |
| Descriptor | 80th Percentile | 90th Percentile | 95th Percentile | Maximum | Minimum | Average |
|---|---|---|---|---|---|---|
| MW (Da) | 408.4 | 447.2 | 503.3 | 2664.0 | 42.0 | 319.5 |
| TPSA (Å2) | 97.2 | 126.6 | 157.0 | 922.0 | 9.0 | 80.0 |
| NRB [a] | 7 | 8 | 10 | 30 | 2 | 4 |
| LogP | 4.8 | 5.7 | 6.1 | 8.3 | −5.0 | 3.0 |
| HBA [b] | 7 | 9 | 10 | 42 | 0 | 5 |
| HBD [c] | 2 | 2 | 3 | 30 | 0 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yáñez, O.; Alegría-Arcos, M.; Suardiaz, R.; Morales-Quintana, L.; Castro, R.I.; Palma-Olate, J.; Galarza, C.; Catagua-González, Á.; Rojas-Pérez, V.; Urra, G.; et al. Calcium-Alginate-Chitosan Nanoparticle as a Potential Solution for Pesticide Removal, a Computational Approach. Polymers 2023, 15, 3020. https://doi.org/10.3390/polym15143020
Yáñez O, Alegría-Arcos M, Suardiaz R, Morales-Quintana L, Castro RI, Palma-Olate J, Galarza C, Catagua-González Á, Rojas-Pérez V, Urra G, et al. Calcium-Alginate-Chitosan Nanoparticle as a Potential Solution for Pesticide Removal, a Computational Approach. Polymers. 2023; 15(14):3020. https://doi.org/10.3390/polym15143020
Chicago/Turabian StyleYáñez, Osvaldo, Melissa Alegría-Arcos, Reynier Suardiaz, Luis Morales-Quintana, Ricardo I. Castro, Jonathan Palma-Olate, Christian Galarza, Ángel Catagua-González, Víctor Rojas-Pérez, Gabriela Urra, and et al. 2023. "Calcium-Alginate-Chitosan Nanoparticle as a Potential Solution for Pesticide Removal, a Computational Approach" Polymers 15, no. 14: 3020. https://doi.org/10.3390/polym15143020