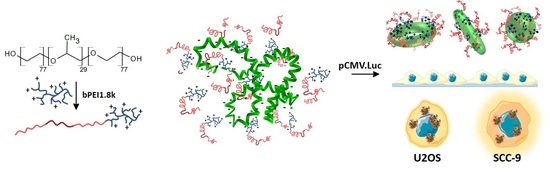
Novel Non-Viral Vectors Based on Pluronic® F68PEI with Application in Oncology Field
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis and Characterization of Copolymer F68PEI
2.3. TNBS Assay
2.4. Buffering Capacity of F68PEI
2.5. Preparation of Polyplexes
2.6. Physicochemical Characterization of F68PEI Nanosystems
2.7. Gel Retardation Assay
2.8. Structural Stability of Polyplexes
2.9. Cell Culture
2.10. Cell Viability
2.11. Transfection Activity
2.12. Statistical Analysis
3. Results and Discussion
3.1. Copolymer Synthesis and Characterization
3.2. Buffering Capacity of F68PEI
3.3. Polyplex Preparation and Characterization
3.4. Cytotoxicity of F68 Polyplexes
3.5. Transfection Efficiency of F68 Polyplexes
3.6. Physicochemical Characterization of F68 Polyplexes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jarak, I.; Pereira-Silva, M.; Paiva-Santos, A.C.; Veiga, F.; Cabral, H.; Figueiras, A. Multifunctional polymeric micelle-based nucleic acid delivery: Current advances and future perspectives. Appl. Mater. Today 2021, 25, 101217. [Google Scholar] [CrossRef]
- Jarak, I.; Varela, C.L.; Tavares da Silva, E.; Roleira, F.F.M.; Veiga, F.; Figueiras, A. Pluronic-based nanovehicles: Recent advances in anticancer therapeutic applications. Eur. J. Med. Chem. 2020, 206, 112526. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, A.; Xu, Z.; Ingram, N.; Coletta, P.L.; Millner, P.A.; Tyler, A.I.I.; Hughes, T.A. Hyaluronic-Acid-Tagged Cubosomes Deliver Cytotoxics Specifically to CD44-Positive Cancer Cells. Mol. Pharm. 2022. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Alakhova, D.Y.; Kabanov, A.V. Pluronics and MDR reversal: An update. Mol. Pharm. 2014, 11, 2566–2578. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Yin, Q.; Chen, L.; Zhang, Z.; Li, Y. Co-delivery of paclitaxel and survivin shRNA by pluronic P85-PEI/TPGS complex nanoparticles to overcome drug resistance in lung cancer. Biomaterials 2012, 33, 8613–8624. [Google Scholar] [CrossRef]
- Magalhães, M.; Jorge, J.; Gonçalves, A.C.; Sarmento-Ribeiro, A.B.; Carvalho, R.; Figueiras, A.; Santos, A.C.; Veiga, F. miR-29b and retinoic acid co-delivery: A promising tool to induce a synergistic antitumoral effect in non-small cell lung cancer cells. Drug Deliv. Transl. Res. 2020, 10, 1367–1380. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Chen, Z.; He, Y. Intracellular redox-responsive nanocarrier for plasmid delivery: In vitro characterization and in vivo studies in mice. Int. J. Nanomed. 2016, 11, 5245–5256. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Zhao, W.; Liu, K.; Yu, Q.; Mao, Y.; Lu, Z.; Zhang, Y.; Zhu, M. Low-Molecular Weight Polyethylenimine Modified with Pluronic 123 and RGD- or Chimeric RGD-NLS Peptide: Characteristics and Transfection Efficacy of Their Complexes with Plasmid DNA. Molecules 2016, 21, 655. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Wu, B.; Tucker, J.D.; Lu, P.; Lu, Q. Poly(ester amine) constructed from polyethylenimine and pluronic for gene delivery in vitro and in vivo. Drug Deliv. 2016, 23, 3224–3233. [Google Scholar] [CrossRef] [Green Version]
- Vinogradov, S.V.; Batrakova, E.V.; Li, S.; Kabanov, A.V. Mixed polymer micelles of amphiphilic and cationic copolymers for delivery of antisense oligonucleotides. J. Drug Target. 2004, 12, 517–526. [Google Scholar] [CrossRef]
- Liang, W.; Gong, H.; Yin, D.; Lu, S.; Fu, Q. High-molecular-weight polyethyleneimine conjuncted pluronic for gene transfer agents. Chem. Pharm. Bull. 2011, 59, 1094–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, W.; Wu, X.; Ding, B.; Gao, J.; Cai, Z.; Zhang, W.; Yin, D.; Wang, X.; Zhu, Q.; Liu, J.; et al. Degradable gene delivery systems based on Pluronics-modified low-molecular-weight polyethylenimine: Preparation, characterization, intracellular trafficking, and cellular distribution. Int. J. Nanomed. 2012, 7, 1127–1138. [Google Scholar] [CrossRef] [Green Version]
- Sanjula, B.; Shah, F.M.; Javed, A.; Alka, A. Effect of poloxamer 188 on lymphatic uptake of carvedilol-loaded solid lipid nanoparticles for bioavailability enhancement. J. Drug Target. 2009, 17, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Grapentin, C.; Müller, C.; Kishore, R.; Adler, M.; ElBialy, I.; Friess, W.; Huwyler, J.; Khan, T. Protein-Polydimethylsiloxane Particles in Liquid Vial Monoclonal Antibody Formulations Containing Poloxamer 188. J. Pharm. Sci. 2020, 109, 2393–2404. [Google Scholar] [CrossRef]
- Bollenbach, L.; Buske, J.; Mäder, K.; Garidel, P. Poloxamer 188 as surfactant in biological formulations—An alternative for polysorbate 20/80? Int. J. Pharm. 2022, 620, 121706. [Google Scholar] [CrossRef]
- Saqafi, B.; Rahbarizadeh, F. Effect of PEI surface modification with PEG on cytotoxicity and transfection efficiency. Micro Nano Lett. 2018, 13, 1090–1095. [Google Scholar] [CrossRef]
- Cordeiro, R.A.; Mendonça, P.V.; Coelho, J.; Faneca, H. Engineering silica-polymer hybrid nanosystems for dual drug and gene delivery. Biomater. Adv. 2022, 135, 212742. [Google Scholar] [CrossRef]
- Arranja, A.; Ivashchenko, O.; Denkova, A.G.; Morawska, K.; van Vlierberghe, S.; Dubruel, P.; Waton, G.; Beekman, F.J.; Schosseler, F.; Mendes, E. SPECT/CT Imaging of Pluronic Nanocarriers with Varying Poly(ethylene oxide) Block Length and Aggregation State. Mol. Pharm. 2016, 13, 1158–1165. [Google Scholar] [CrossRef] [Green Version]
- Degors, I.M.S.; Wang, C.; Rehman, Z.U.; Zuhorn, I.S. Carriers Break Barriers in Drug Delivery: Endocytosis and Endosomal Escape of Gene Delivery Vectors. Acc. Chem. Res. 2019, 52, 1750–1760. [Google Scholar] [CrossRef] [Green Version]
- Benjaminsen, R.V.; Mattebjerg, M.A.; Henriksen, J.R.; Moghimi, S.M.; Andresen, T.L. The possible “proton sponge” effect of polyethylenimine (PEI) does not include change in lysosomal pH. Mol. Ther. J. Am. Soc. Gene Ther. 2013, 21, 149–157. [Google Scholar] [CrossRef]
- Sabin, J.; Alatorre-Meda, M.; Miñones, J.; Domínguez-Arca, V.; Prieto, G. New insights on the mechanism of polyethylenimine transfection and their implications on gene therapy and DNA vaccines. Colloids Surf. B Biointerfaces 2022, 210, 112219. [Google Scholar] [CrossRef] [PubMed]
- Banquy, X.; Lee, D.W.; Kristiansen, K.; Gebbie, M.A.; Israelachvili, J.N. Interaction Forces between Supported Lipid Bilayers in the Presence of PEGylated Polymers. Biomacromolecules 2016, 17, 88–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelat, P.B.; Plant, L.D.; Wang, J.C.; Lee, E.; Marks, J.D. The membrane-active tri-block copolymer pluronic F-68 profoundly rescues rat hippocampal neurons from oxygen-glucose deprivation-induced death through early inhibition of apoptosis. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 12287–12299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahay, G.; Batrakova, E.V.; Kabanov, A.V. Different internalization pathways of polymeric micelles and unimers and their effects on vesicular transport. Bioconjug. Chem. 2008, 19, 2023–2029. [Google Scholar] [CrossRef] [Green Version]
- Zhirnov, A.E.; Demina, T.V.; Krylova, O.O.; Grozdova, I.D.; Melik-Nubarov, N.S. Lipid composition determines interaction of liposome membranes with Pluronic L61. Biochim. Et Biophys. Acta (BBA)—Biomembr. 2005, 1720, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Rey-Rico, A.; Cucchiarini, M. PEO-PPO-PEO Tri-Block Copolymers for Gene Delivery Applications in Human Regenerative Medicine—An Overview. Int. J. Mol. Sci. 2018, 19, 775. [Google Scholar] [CrossRef] [Green Version]
- Kuo, J.H. Effect of Pluronic-block copolymers on the reduction of serum-mediated inhibition of gene transfer of polyethyleneimine-DNA complexes. Biotechnol. Appl. Biochem. 2003, 37, 267–271. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, L.; Li, Y. Addition of pluronics® to reducible disulfide-bond-containing Pluronic®-PEI-SS specifically enhances circulation time in vivo and transfection efficiency in vitro. J. Biomed. Mater. Research. Part B Appl. Biomater. 2014, 102, 1268–1276. [Google Scholar] [CrossRef]
- Wang, M.; Lu, P.; Wu, B.; Tucker, J.D.; Cloer, C.; Lu, Q. High efficiency and low toxicity of polyethyleneimine modified Pluronics (PEI–Pluronic) as gene delivery carriers in cell culture and dystrophic mdx mice. J. Mater. Chem. 2012, 22, 6038–6046. [Google Scholar] [CrossRef]
- Alakhova, D.Y.; Rapoport, N.Y.; Batrakova, E.V.; Timoshin, A.A.; Li, S.; Nicholls, D.; Alakhov, V.Y.; Kabanov, A.V. Differential metabolic responses to pluronic in MDR and non-MDR cells: A novel pathway for chemosensitization of drug resistant cancers. J. Control. Release Off. J. Control. Release Soc. 2010, 142, 89–100. [Google Scholar] [CrossRef]
- Batrakova, E.V.; Li, S.; Alakhov, V.Y.; Miller, D.W.; Kabanov, A.V. Optimal structure requirements for pluronic block copolymers in modifying P-glycoprotein drug efflux transporter activity in bovine brain microvessel endothelial cells. J. Pharmacol. Exp. Ther. 2003, 304, 845–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Li, Y.; Li, C.; Yi, X.; Wang, R.; Lee, S.M.-Y.; Zheng, Y. Pluronic P85/F68 Micelles of Baicalein Could Interfere with Mitochondria to Overcome MRP2-Mediated Efflux and Offer Improved Anti-Parkinsonian Activity. Mol. Pharm. 2017, 14, 3331–3342. [Google Scholar] [CrossRef]
- Aydin, S.; YalvaÇ, M.; Ozcan, F.; Sahin, F. Pluronic PF68 increases transfection efficiency in electroporationof mesenchymal stem cells. Turk. J. Biol. 2016, 40, 747–754. [Google Scholar] [CrossRef]
- Samith, V.D.; Miño, G.; Ramos-Moore, E.; Arancibia-Miranda, N. Effects of pluronic F68 micellization on the viability of neuronal cells in culture. J. Appl. Polym. Sci. 2013, 130, 2159–2164. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.K.; Lemieux, P.; Vinogradov, S.V.; Gebhart, C.L.; Guérin, N.; Paradis, G.; Bronich, T.K.; Alakhov, V.Y.; Kabanov, A.V. Evaluation of polyether-polyethyleneimine graft copolymers as gene transfer agents. Gene Ther. 2000, 7, 126–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Tian, Q.; Huang, Z.; Fan, D.; She, Z.; Liu, X.; Cheng, X.; Yu, B.; Deng, Y. Self-assembled micelles of novel amphiphilic copolymer cholesterol-coupled F68 containing cabazitaxel as a drug delivery system. Int. J. Nanomed. 2014, 9, 2307–2317. [Google Scholar] [CrossRef] [Green Version]
- Pezzoli, D.; Giupponi, E.; Mantovani, D.; Candiani, G. Size matters for in vitro gene delivery: Investigating the relationships among complexation protocol, transfection medium, size and sedimentation. Sci. Rep. 2017, 7, 44134. [Google Scholar] [CrossRef] [PubMed]






| N/P | Particle Size (nm) | PDI | Zeta Potential (mV) | N/P | Particle Size (nm) | PDI | Zeta Potential (mV) | ||
|---|---|---|---|---|---|---|---|---|---|
| F68PEI | 5 | 210.5 | 0.41 | 51.4 | F68PEI/F68 1:5 | 5 | 151.4 | 0.57 | 43.3 |
| 10 | 273.0 (591.3) * | 0.52 (0.62) | 41.3 (13.6) | 10 | 188.6 (883.1) | 0.50 (0.68) | 40.4 (25.3) | ||
| 25 | 310.7 (486.1) | 0.53 (0.52) | 22.1 (35.3) | 25 | 113.6 (582.6) | 0.54 (0.68) | 22.8 (31.6) | ||
| 50 | 277.0 (475.0) | 0.44 (0.54) | 41.3 (43.9) | 50 | 183.4 (248.2) | 0.39 (0.46) | 20.1 (40.2) | ||
| 75 | 384.0 | 0.73 | 36.9 | 75 | 86.6 | 0.56 | 25.1 | ||
| 100 | 217.4 | 0.49 | 40.4 | 100 | 101.1 | 0.55 | 28.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, I.; Domingues, C.; Jarak, I.; Carvalho, R.A.; Cordeiro, R.A.; Dourado, M.; Veiga, F.; Faneca, H.; Figueiras, A. Novel Non-Viral Vectors Based on Pluronic® F68PEI with Application in Oncology Field. Polymers 2022, 14, 5315. https://doi.org/10.3390/polym14235315
Silva I, Domingues C, Jarak I, Carvalho RA, Cordeiro RA, Dourado M, Veiga F, Faneca H, Figueiras A. Novel Non-Viral Vectors Based on Pluronic® F68PEI with Application in Oncology Field. Polymers. 2022; 14(23):5315. https://doi.org/10.3390/polym14235315
Chicago/Turabian StyleSilva, Inês, Cátia Domingues, Ivana Jarak, Rui A. Carvalho, Rosemeyre A. Cordeiro, Marília Dourado, Francisco Veiga, Henrique Faneca, and Ana Figueiras. 2022. "Novel Non-Viral Vectors Based on Pluronic® F68PEI with Application in Oncology Field" Polymers 14, no. 23: 5315. https://doi.org/10.3390/polym14235315