Synthesis and Comparative Study of Nanoparticles Derived from Bovine and Human Serum Albumins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
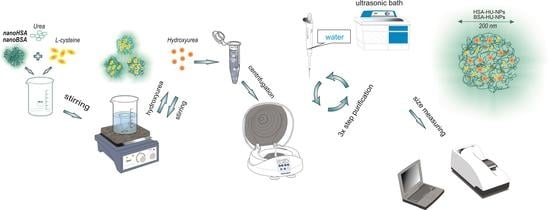
2.2. Preparation of Drug-Free Nanoparticles Derived from BSA and HSA
2.3. Preparation of Proteinaceous Nanoparticles Loaded with Hydroxyurea
- (i)
- The process of serum albumin-derived nanoparticles, loading with hydroxyurea (HU) through the adsorption, consisted of the synthesis of the drug-free nanoparticles (Section 2.2) and their subsequent loading with the drug. For this aim, the hydroxyurea was added to 22 mL of the nanoparticles’ aqueous suspension (HU concentration was either 2, or 4, or 6 mg/mL) in an amount of either 45 or 90, or 135 mg. The resulting suspensions were stirred at a constant rate of 200 rpm for 2 h. The final stage was the centrifugation of the suspensions in three cycles at 14,000 rpm to remove the unbound HU.
- (ii)
- The protein nanoparticles have been fabricated essentially in the same way as described above in Section 2.2, except for after completion of the desolvating process, 3 mL of aqueous HU solution (2 mg/mL) was added to the nanoparticulate dispersion followed by the system treatment with urea and L-cystein mixture under the conditions analogous to those in Section 2.2.
2.4. Size Measurement, Fourier-Transform Infrared Spectroscopy, Zeta Potential Analysis, Surface Morphology and Thermal Analysis of Serum Albumin-Derived Nanoparticles
2.5. Yield and Drug Loading Efficiency of BSA(HSA)-NPs
2.6. Determination of the In Vitro HU Release
2.7. Statistical Analysis
3. Results and Discussion
3.1. Optimization of Nanoparticles Preparation
3.2. Loading of Serum Albumin Nanoparticles with Hydroxyurea
3.3. Physicochemical Characteristics of BSA-NPs with Immobilized Hydroxyurea
3.4. In Vitro Drug Release
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kreuter, J. Colloidal Drug Delivery Systems; Marcel Dekker: New York, NY, USA, 1994; pp. 66–344. [Google Scholar]
- Marty, J.J.; Oppenheim, R.C.; Speiser, P. Nanoparticles—A new colloidal drug delivery system. Pharm. Acta Helv. 1978, 53, 17–23. [Google Scholar]
- Peters, T. All about Albumin: Biochemistry, Genetics, and Medical Application; Academic Press: Cambridge, MA, USA, 1995; p. 432. [Google Scholar]
- Hulzebos, C.V.; Dijk, P.H. Bilirubin–albumin binding, bilirubin/albumin ratios, and free bilirubin levels: Where do we stand? Semin Perinatol. 2014, 38, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Al-Saadi, A.; Yu, C.; Khutoryanskiy, V.; Shih, S.-J.; Crossley, A.; Tsang, S. Layer-by-layer electrostatic entrapment of protein molecules on superparamagnetic nanoparticle: New strategy to enhance adsorption capacity and maintain biological activity. J. Phys. Chem. C. 2009, 113, 15260–15265. [Google Scholar] [CrossRef]
- Fiume, L.; Busi, C.; DiStefano, G.; Mattioli, A. Coupling of antiviral nucleoside analogs to lactosaminated human albumin by using the imidazolides of their phosphoric esters. Anal. Biochem. 1993, 212, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Warnecke, A.; Fichtner, I.; Garmenn, D.; Jaehde, U.; Kratz, F. Synthesis and biological activity of water-soluble maleimide derivative of the anticancer drug carboplatin designed as albumin-binding prodrugs. Bioconjug. Chem. 2004, 15, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Osborn, B.L.; Olsen, H.S.; Nardelli, B.; Murray, J.H.; Zhou, J.X.; Garcia, A.; Moody, G.; Zaritskaya, L.S.; Sung, C. Pharmacokinetic and pharmacodynamic studies of a human serum albumin-interferon-alpha fusion protein in cynomolgus monkeys. J. Pharmacol. Exp. Ther. 2002, 303, 540–548. [Google Scholar] [CrossRef] [Green Version]
- Kratz, F. A clinical update of using albumin as a drug vehicle—A commentary. J. Control. Release 2014, 190, 331–336. [Google Scholar] [CrossRef]
- Chen, Q.; Liang, C.; Wang, C.; Liu, Z. An imagable and photothermal “Abraxane-like” nanodrug for combination cancer therapy to treat subcutaneous and metastatic breast tumors. Adv. Mater. 2015, 27, 903–910. [Google Scholar] [CrossRef]
- Bertrand, N.; Wu, J.; Xu, X.; Kamaly, N.; Farokhzad, O.C. Cancer nanotechnology: The impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Deliv. Rev. 2014, 66, 2–25. [Google Scholar] [CrossRef] [Green Version]
- Miele, E.; Spinelli, G.P.; Miele, E.; Tomao, F.; Tomao, S. Albumin-bound formulation of paclitaxel (Abraxane (R) ABI-007) in the treatment of breast cancer. Int. J. Nanomed. 2009, 4, 99–105. [Google Scholar]
- Hawkins, M.J.; Soon-Shiong, P.; Desai, N. Protein nanoparticles as drug carriers in clinical medicine. Adv. Drug Deliv. Rev. 2008, 60, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Luppi, B.; Bigucci, F.; Corace, G.; Delucca, A.; Cerchiara, T.; Sorrenti, M.; Catenacci, L.; Di Pietra, A.; Zecchi, V. Albumin nanoparticles carrying cyclodextrins for nasal delivery of the anti-Alzheimer drug tacrine. Eur. J. Pharm. Sci. 2011, 44, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Sebak, S.; Mirzael, M.; Malhotra, M.; Kulamarva, A.; Prakash, S. Human serum albumin nanoparticles as an efficient noscapine drug delivery system for potential use in breast cancer: Preparation and in vitro analysis. Int. J. Nanomed. 2010, 5, 525–532. [Google Scholar]
- Wartlick, H.; Spänkuch-Schmitt, B.; Strebhardt, K.; Kreuter, J.; Langer, K. Tumour cell delivery of antisense oligonucleotides by human serum albumin nanoparticles. J. Control. Release 2004, 18, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Tazhbayev, Y.; Mukashev, O.; Burkeev, M.; Kreuter, J. Hydroxyurea-Loaded Albumin Nanoparticles: Preparation, Characterization, and In Vitro Studies. Pharmaceutics 2019, 11, 410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubino, O.P.; Kowalsky, R.; Swarbrick, J. Albumin microspheres as a drug delivery system: Relation among turbidity ratio, degree of cross-linking, and drug release. Pharm. Res. 1993, 10, 1059–1065. [Google Scholar] [CrossRef]
- Weber, C.; Kreuter, J.; Langer, K. Desolvation process and surface characteristics of HSA-nanoparticles. Int. J. Pharm. 2000, 196, 197–200. [Google Scholar] [CrossRef]
- Mondal, S.; Li, C.; Wang, K. Bovine Serum Albumin Adsorption on Gluteraldehyde Cross-Linked Chitosan Hydrogels. J. Chem. Eng. Data. 2015, 60, 2356–2362. [Google Scholar] [CrossRef]
- Ezpeleta, I.; Irache, J.M.; Gueguen, J.; Orecchioni, A.M. Properties of glutaraldehyde cross-linked vicilin nano- and microparticles. J. Microencapsul. 1997, 14, 557–565. [Google Scholar] [CrossRef]
- Merodio, M.; Arnedo, A.; Renedo, M.J.; Irache, J.M. Ganciclovir-loaded albumin nanoparticles: Characterization and in vitro release properties. Eur. J. Pharm. Sci. 2001, 12, 251–259. [Google Scholar] [CrossRef]
- Li, F.Q.; Su, H.; Wang, J.; Liu, J.Y.; Zhu, Q.G.; Fei, Y.B.; Pan, Y.H.; Hu, J.H. Preparation and characterization of sodium ferulate entrapped bovine serum albumin nanoparticles for liver targeting. Int. J. Pharm. 2008, 349, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhang, H.; Li, X.; Lei, X.; Li, C.; Yin, D.; Fan, X.; Zhang, Q. Synthesis of BSA/Fe3O4 magnetic composite microspheres for adsorption of antibiotics. Mater. Sci. Eng. C Mater. Biol. Appl. 2013, 33, 4401–4408. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Willmott, N.; Anderson, J.; Florence, A.T. Comparison of albumin and casein microspheres as a carrier for doxorubicin. J. Pharm. Pharmacol. 1987, 39, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Sakamoto, H.; Kako, Y. Cross-linking of Caseins by Colloidal Calcium Phosphate in the Presence of Urea. Int. Dairy J. 1991, 1, 67–75. [Google Scholar] [CrossRef]
- Müller, B.G.; Leuenberger, H.; Kissel, T. Albumin nanospheres as carriers for passive drug targeting: An optimized manufacturing technique. Pharm. Res. 1996, 13, 32–37. [Google Scholar] [CrossRef]
- Loiko, O.; Herk, A.M.; Ali, S.; Burkeev, M.; Tazhbayev, Y.; Zhaparova, L. Controlled release of Capreomycin sulfate from pH-responsive nanocapsules. e-Polymers 2013, 13, 1–5. [Google Scholar] [CrossRef]
- Leggio, C.; Galantini, L.; Konarev, P.V.; Pavel, N.V. Urea-Induced Denaturation Process on Defatted Human Serum Albumin and in the Presence of Palmitic Acid. J. Phys. Chem. B. 2009, 113, 12590–12602. [Google Scholar] [CrossRef]
- Zhunuspayev, D.E.; Mun, G.A.; Khutoryanskiy, V.V. Temperature-responsive properties and drug solubilization capacity of amphiphilic copolymers based on N-vinylpyrrolidone and vinyl propyl ether. Langmuir 2010, 26, 7590–7597. [Google Scholar] [CrossRef]
- Lozinsky, V.I.; Okay, O. Basic principles of cryotropic gelation. Adv. Polym. Sci. 2014, 263, 49–102. [Google Scholar]
- Rodionov, I.A.; Grinberg, N.V.; Burova, T.V.; Grinberg, V.; Lozinsky, V.I. Cryostructuring of Polymer Systems. Proteinaceous Wide-Pore Cryogels Generated by the Action of Denaturant/Reductant Mixtures on Bovine Serum Albumin in Moderately-Frozen Aqueous Media. Soft Matter 2015, 11, 4921–4931. [Google Scholar] [CrossRef]
- Lomis, N.; Westfall, S.; Farahdel, L. Human Serum Albumin Nanoparticles for Use in Cancer Drug Delivery: Process Optimization and in vitro Characterization. Nanomaterials 2016, 6, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burkeev, M.Z.; Kreuter, J.; Tazhbayev, Y.M.; Zhaparova, L.Z.; Zhumalieva, T.S. Preparation, characterization and investigation of in vitro release of anti-tuberculosis drug p-amino salicylic acid based on human serum albumin. Bull. Karaganda Univ. 2017, 87, 38–44. [Google Scholar] [CrossRef]
- Akbari, A.; Akbarzadeh, A.; Tehrani, M.; Cohan, R.; Chiani, M.; Mehrabi, M. Development and Characterization of Nanoliposomal Hydroxyurea Against BT-474 Breast Cancer Cells. Adv. Pharm. Bull. 2019, 10, 39–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kufleitner, J.; Wagner, S.; Worek, F.; von Briesen, H.; Kreuter, J. Adsorption of obidoxime onto human serum albumin nanoparticles: Drug loading, particle size and drug release. J. Microencapsul. 2010, 27, 506–513. [Google Scholar] [CrossRef]
- Dreis, S.; Rothweiler, F.; Michaelis, M.; Cinatl, J., Jr.; Kreuter, J.; Langer, K. Preparation, characterisation and maintenance of drug efficacy of doxorubicin-loaded human serum albumin (HSA) nanoparticles. Int. J. Pharm. 2007, 341, 207–214. [Google Scholar] [CrossRef]
- Gao, S.; Sun, J.; Fu, D.; Zhao, H.; Lan, M.; Gao, F. Preparation, characterization and pharmacokinetic studies of tacrolimus-dimethyl-beta-cyclodextrin inclusion complex-loaded albumin nanoparticles. Int. J. Pharm. 2012, 427, 410–416. [Google Scholar] [CrossRef]
- Qu, N.; Lee, J.R.; Sun, Y. Cabazitaxel-loaded human serum albumin nanoparticles as a therapeutic agent against prostate cancer. Int. J. Nanomed. 2016, 11, 3451–3459. [Google Scholar]
- Zhou, W.; Cao, Y.; Sui, D.; Guan, W.; Lu, C.; Xie, J. Ultrastable BSA-capped gold nanoclusters with a polymer-like shielding layer against reactive oxygen species in living cells. Nanoscale 2016, 8, 9614–9620. [Google Scholar] [CrossRef]
- Lunghi, A.; Aloni, C.; Gigante, L.; Mazzei, N.; Cardillo, P. Hydroxyurea explosion: A thermoanalytical and calorimetric study. J. Loss Prev. Proc. Ind. 2002, 15, 489–495. [Google Scholar] [CrossRef]
- Rowe, P. Hydroxyurea in Sickle Cell Disease: Drug Review. Lancet 1995, 345, 311. [Google Scholar] [CrossRef]
- Relkin, P. Thermal unfolding of beta-lactoglobulin, alpha-lactalbumin, and bovine serum albumin. A thermodynamic approach. Crit. Rev. Food Sci. Nutr. 1996, 36, 565–601. [Google Scholar] [CrossRef] [PubMed]
- Michnik, A.; Michalik, K.; Kluczewska, A.; Drzazga, Z. Comparative DSC study of human and bovine serum albumin. J. Therm. Anal. Calorim. 2006, 84, 113–117. [Google Scholar] [CrossRef]









| Type of Initial Albumin | Initial Protein Concentration mg/mL | Particles Size ± SD (nm) | PI ± SD | Zeta-Potential (mV) ±SD | Encapsulation Efficiency (%) ± SD | |||
|---|---|---|---|---|---|---|---|---|
| Before HU Adsorption | After HU Adsorption | Before HU Adsorption | After HU Adsorption | Before HU Adsorption | After HU Adsorption | |||
| BSA | 10 | 157 ± 2 | 175 ± 3 | 0.093 ± 0.010 | 0.181 ± 0.010 | −23 ± 1 | −24 ± 1 | 74 ± 2 |
| 20 | 163 ± 3 | 178 ± 5 | 0.122 ± 0.015 | 0.199 ± 0.010 | −26 ± 1 | −27 ± 1 | 73 ± 2 | |
| 30 | 167 ± 5 | 183 ± 6 | 0.202 ± 0.020 | 0.227 ± 0.010 | −25 ± 1 | −23 ± 1 | 70 ± 2 | |
| HSA | 30 | 216 ± 9 | 277 ± 2 | 0.148 ± 0.015 | 0.200 ± 0.010 | −24 ± 1 | −22 ± 0 | 68 ± 2 |
| Type of Initial Albumin, mg/mL | Particles Size ± SD (nm) | PI ± SD | Zeta-Potential (mV) ± SD | Nanoparticle Yield, Protein % | Encapsulation Efficiency ± SD |
|---|---|---|---|---|---|
| BSA (2 mg/mL) | 239 ± 10 | 0.242 ± 0.015 | −20 ± 3 | 100% | 78 ± 1 |
| HSA (2 mg/mL) | 330 ± 12 | 0.287 ± 0.010 | −17 ± 2 | 100% | 69 ± 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tazhbayev, Y.; Mukashev, O.; Burkeyev, M.; Lozinsky, V.I. Synthesis and Comparative Study of Nanoparticles Derived from Bovine and Human Serum Albumins. Polymers 2020, 12, 1301. https://doi.org/10.3390/polym12061301
Tazhbayev Y, Mukashev O, Burkeyev M, Lozinsky VI. Synthesis and Comparative Study of Nanoparticles Derived from Bovine and Human Serum Albumins. Polymers. 2020; 12(6):1301. https://doi.org/10.3390/polym12061301
Chicago/Turabian StyleTazhbayev, Yerkeblan, Olzhas Mukashev, Meiram Burkeyev, and Vladimir I. Lozinsky. 2020. "Synthesis and Comparative Study of Nanoparticles Derived from Bovine and Human Serum Albumins" Polymers 12, no. 6: 1301. https://doi.org/10.3390/polym12061301