Physicochemical Characterization and Immunomodulatory Activity of a Novel Acid Polysaccharide from Solanum muricatum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
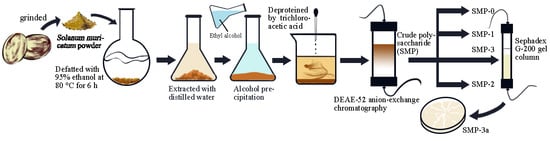
2.2. Extraction of Crude Polysaccharide (SMP)
2.3. Purification of SMP
2.4. FT-IR Spectrum Analysis
2.5. Determination of Molecular Weight
2.6. Reduction of Carboxyl Groups
2.7. Chemical Components and Monosaccharide Composition Analysis
2.8. Methylation Analysis of SMP-3a-R
2.9. NMR Analysis
2.10. Scanning Electron Microscopy (SEM) Observation
2.11. Thermal Gravimetric Analysis (TGA) and Differential Scanning Calorimetric (DSC) Analysis
2.12. Immunomodulatory Activity of SMP-3a
2.12.1. Cell Culture
2.12.2. RAW264.7 Macrophage Proliferation Assay
2.12.3. Measurement of Cytokine Production
2.13. Statistical Analysis
3. Results and Discussion
3.1. Purification of SMP
3.2. FT-IR Spectroscopy Analysis
3.3. Molecular Weight of SMP-3a
3.4. Chemical Components and Monosaccharide Composition
3.5. Methylation Analysis of SMP-3a
3.6. NMR Analysis of SMP-3a

3.7. Morphological Analysis
3.8. Thermal Properties
3.9. Immunomodulatory Activity of SMP-3a
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kumar, A.; Adak, T.; Rajan, S. Pepino (Solanum muricatum Ait.): A potential future crop for subtropics. Trop. Plant Res. 2017, 4, 514–517. [Google Scholar] [CrossRef]
- Martínez-Romero, D.; Serrano, M.; Valero, D. Physiological changes in pepino (Solanum muricatum Ait.) fruit stored at chilling and non-chilling temperatures. Postharvest Biol. Technol. 2003, 30, 177–186. [Google Scholar] [CrossRef]
- Kola, O.; Simsek, M.; Duran, H.; Bozkir, H. HPLC determination of carotenoid, organic acid, and sugar content in pepino (Solanum muricatum) fruit during the ripening period. Chem. Nat. Compd. 2015, 51, 132–135. [Google Scholar] [CrossRef]
- Shathish, K.; Guruvayoorappan, C. Solanum muricatum Ait. inhibits inflammation and cancer by modulating the immune system. J. Cancer Res. Ther. 2014, 10, 623–630. [Google Scholar]
- Prohens, J.; Nuez, F.; Kollmannsberger, H.; Nitz, S.; Rodríguez-Burruezo, A. Analysis of the Volatile Aroma Constituents of Parental and Hybrid Clones of Pepino (Solanum muricatum). J. Agric. Food Chem. 2007, 52, 5663–5669. [Google Scholar]
- Hsu, C.C.; Guo, Y.R.; Wang, Z.H.; Yin, M.C. Protective effects of an aqueous extract from pepino (Solanum muricatum Ait.) in diabetic mice. J. Sci. Food Agric. 2011, 91, 1517–1522. [Google Scholar] [CrossRef]
- Yu, Y.; Shen, M.; Song, Q.; Xie, J. Biological activities and pharmaceutical applications of polysaccharide from natural resources: A review. Carbohydr. Polym. 2018, 183, 91–101. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, P.; Zhao, S.; Nie, C.; Wang, N.; Du, X.; Zhou, Y. Characterization, antioxidant activity and immunomodulatory activity of polysaccharides from the swollen culms of Zizania latifolia. Int. J. Biol. Macromol. 2017, 95, 809–817. [Google Scholar] [CrossRef]
- Li, J.E.; Nie, S.P.; Xie, M.Y.; Li, C. Isolation and partial characterization of a neutral polysaccharide from Mosla chinensis Maxim. cv. Jiangxiangru and its antioxidant and immunomodulatory activities. J. Funct. Foods 2014, 6, 410–418. [Google Scholar] [CrossRef]
- Seedevi, P.; Moovendhan, M.; Viramani, S.; Shanmugam, A. Bioactive potential and structural chracterization of sulfated polysaccharide from seaweed (Gracilaria corticata). Carbohydr. Polym. 2017, 155, 516–524. [Google Scholar] [CrossRef]
- Ding, X.; Zhu, F.; Gao, S. Purification, antitumour and immunomodulatory activity of water-extractable and alkali-extractable polysaccharides from Solanum nigrum L. Food Chem. 2012, 131, 677–684. [Google Scholar] [CrossRef]
- Zhu, K.; Zhang, Y.; Nie, S.; Xu, F.; He, S.; Gong, D.; Wu, G.; Tan, L. Physicochemical properties and in vitro antioxidant activities of polysaccharide from Artocarpus heterophyllus Lam. pulp. Carbohydr. Polym. 2017, 155, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhang, J.; Chen, F.; Chen, X.; Zhou, Z.; Wang, H. Activation of RAW264.7 macrophages by the polysaccharide from the roots of Actinidia eriantha and its molecular mechanisms. Carbohydr. Polym. 2015, 121, 388–402. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Chen, J.; Tian, S.; Gu, H.; Li, N.; Sun, Y.; Ru, J.; Wang, J. Extraction optimization, preliminary characterization and immunological activities in vitro of polysaccharides from Elaeagnus angustifolia L. pulp. Carbohydr. Polym. 2016, 151, 348–357. [Google Scholar] [CrossRef] [PubMed]
- DuBois, M.; Smith, F.; Rebers, P.A.; Gilles, K.A.; Hamilton, J.K. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Liu, Y.; Su, P.; Xu, J.; Chen, S.; Zhang, J.; Zhou, S.; Wang, Y.; Tang, Q.; Wang, Y. Structural characterization of a bioactive water-soluble heteropolysaccharide from Nostoc sphaeroids kütz. Carbohydr. Polym. 2018, 200, 552–559. [Google Scholar] [CrossRef]
- Taylorf, R.L.; Conrad, H.E. Stoichiometric Depolymerization of Polyuronides and Glycosaminoglycuronans to Monosaccharides following Reduction of Their Carbodiimide-Activated Carboxyl Groups. Biochemistry 1972, 11, 1383–1388. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Blumenkrantz, N.; Asboe-Hansen, G. New method for quantitative determination of uronic acids. Anal. Biochem. 1973, 54, 484–489. [Google Scholar] [CrossRef]
- Zhang, H.; Li, W.J.; Nie, S.P.; Chen, Y.; Wang, Y.X.; Xie, M.Y. Structural characterisation of a novel bioactive polysaccharide from Ganoderma atrum. Carbohydr. Polym. 2012, 88, 1047–1054. [Google Scholar] [CrossRef]
- Chen, M.M.; Wu, J.; Shi, S.; Chen, Y.; Wang, H.; Fan, H.; Wang, S. Structure analysis of a heteropolysaccharide from Taraxacum mongolicum Hand.-Mazz. and anticomplementary activity of its sulfated derivatives. Carbohydr. Polym. 2016, 152, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Needs, P.W.; Selvendran, R.R. Avoiding oxidative degradation during sodium hydroxide/methyl iodide-mediated carbohydrate methylation in dimethyl sulfoxide. Carbohydr. Res. 1993, 245, 1–10. [Google Scholar] [CrossRef]
- Chen, Y.; Li, X.H.; Zhou, L.Y.; Li, W.; Liu, L.; Wang, D.D.; Zhang, W.N.; Hussain, S.; Tian, X.H.; Lu, Y.M. Structural elucidation of three antioxidative polysaccharides from Tricholoma lobayense. Carbohydr. Polym. 2017, 157, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Nie, C.; Zhu, P.; Ma, S.; Wang, M.; Hu, Y. Purification, characterization and immunomodulatory activity of polysaccharides from stem lettuce. Carbohydr. Polym. 2018, 188, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, B.; Ibrahim, S.A.; Gao, S.S.; Yang, H.; Huang, W. Purification, characterization and antioxidant activity of polysaccharides from Flammulina velutipes residue. Carbohydr. Polym. 2016, 145, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Kong, F.; Ni, H.; Mo, Z.; Wan, J.B.; Hua, D.; Yan, C. Structural characterization, α-glucosidase inhibitory and DPPH scavenging activities of polysaccharides from guava. Carbohydr. Polym. 2016, 144, 106–114. [Google Scholar] [CrossRef]
- Ren, Y.Y.; Zhu, Z.Y.; Sun, H.Q.; Chen, L.J. Structural characterization and inhibition on α-glucosidase activity of acidic polysaccharide from Annona squamosa. Carbohydr. Polym. 2017, 174, 1–12. [Google Scholar] [CrossRef]
- Zhang, Q.; Cheng, M.; Mou, X.; Zhang, X.; Lv, J.; Fan, Q.; Cao, K.; Xu, Y.; Wu, Y. Structure characterization of two functional polysaccharides from Polygonum multiflorum and its immunomodulatory. Int. J. Biol. Macromol. 2018, 113, 195–204. [Google Scholar] [CrossRef]
- Liu, W.; Liu, Y.; Zhu, R.; Yu, J.; Lu, W.; Pan, C.; Yao, W.; Gao, X. Structure characterization, chemical and enzymatic degradation, and chain conformation of an acidic polysaccharide from Lycium barbarum L. Carbohydr. Polym. 2016, 147, 114–124. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, M.; Liu, J.; Yang, Z.; Shang, F. Structural analysis of a homogeneous polysaccharide from Achatina fulica. Int. J. Biol. Macromol. 2017, 98, 786–792. [Google Scholar]
- Xie, J.H.; Zhang, F.; Wang, Z.J.; Shen, M.Y.; Nie, S.P.; Xie, M.Y. Preparation, characterization and antioxidant activities of acetylated polysaccharides from Cyclocarya paliurus leaves. Carbohydr. Polym. 2015, 133, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Qu, H.; Jia, J.; Kuang, C.; Wen, Y.; Yan, H.; Gui, Z. Characterization, antioxidant and antitumor activities of polysaccharides from purple sweet potato. Carbohydr. Polym. 2015, 132, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xue, Y. Purification, chemical characterization and antioxidant activities of a novel polysaccharide from Auricularia polytricha. Int. J. Biol. Macromol. 2018, 120, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.Z.; Meng, M.; Duan, S.Q.; Feng, C.C.; Wang, C.L. Structure characterization, physicochemical property and immunomodulatory activity on RAW264.7 cells of a novel triple-helix polysaccharide from Craterellus cornucopioides. Int. J. Biol. Macromol. 2019, 126, 796–804. [Google Scholar] [CrossRef]




| Retention Time (min) | Methylated Sugar (Alditol Acetate) | Molar Ratios | Type of Linkage | Mass Fragments (m/z) |
|---|---|---|---|---|
| 11.47 | 2,3,5-Me3-Araf | 1 | Araf-(1→ | 71,87,101,117,129,145,161 |
| 17.11 | 2,3-Me2-Araf | 2.08 | →5)-Araf-(1→ | 71,87,99,101,117,129,161,189 |
| 20.45 | 3,4-Me2-Rhap | 1.16 | →2)-Rhap-1→ | 87,101,117,129,143,159,189 |
| 24.23 | 2,3,6-Me3-Galp | 1.13 | →4)-Galp-(1→ | 87,99,101,113,117,129,131,161,173,233 |
| 31.26 | 2,3-Me2-Galp | 1 | →4,6)-Galp-(1→ | 71,85,87,99,101,117,127,159,161,201 |
| Glycosyl Residues | C1 | C2 | C3 | C4 | C5 | C6 | |
|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H4 | H5/H5a | H5b/H6a | H6b | |
| →5)-α-l-Araf-(1→ | 108.81 | 82.11 | 78.05 | 83.61 | 68.20 | ||
| A | 5.00 | 4.04 | 3.92 | 4.12 | 3.79 | 3.72 | |
| →4)-α-d-GalpA-(1→ | 100.55 | 69.49 | 69.98 | 79.04 | 72.08 | 175.13 | |
| B | 4.99 | 3.67 | 3.92 | 4.34 | 4.71 | ||
| →2)-α-l-Rhap-(1→ | 100.07 | 77.70 | 70.70 | 73.22 | 70.58 | 17.83 | |
| C | 5.15 | 4.03 | 3.78 | 3.33 | 3.64 | 1.16 | 1.22 |
| α-l-Araf-(1→ | 110.50 | 83.40 | 77.65 | 79.26 | 62.21 | ||
| D | 5.16 | 4.12 | 4.03 | 4.34 | 3.64 | 3.72 | |
| →4,6)-α-d-Galp-(1→ | 98.94 | 70.80 | 77.60 | 78.40 | 69.12 | 66.80 | |
| E | 4.94 | 3.81 | 4.02 | 4.35 | 3.83 | 3.68 | 3.76 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, H.; Xu, Q.; Li, X.; Elango, J.; Wu, W.; Xu, J. Physicochemical Characterization and Immunomodulatory Activity of a Novel Acid Polysaccharide from Solanum muricatum. Polymers 2019, 11, 1972. https://doi.org/10.3390/polym11121972
Yue H, Xu Q, Li X, Elango J, Wu W, Xu J. Physicochemical Characterization and Immunomodulatory Activity of a Novel Acid Polysaccharide from Solanum muricatum. Polymers. 2019; 11(12):1972. https://doi.org/10.3390/polym11121972
Chicago/Turabian StyleYue, Heng, Qianqian Xu, Xianheng Li, Jeevithan Elango, Wenhui Wu, and Jianfeng Xu. 2019. "Physicochemical Characterization and Immunomodulatory Activity of a Novel Acid Polysaccharide from Solanum muricatum" Polymers 11, no. 12: 1972. https://doi.org/10.3390/polym11121972