Marine PET Hydrolase (PET2): Assessment of Terephthalate- and Indole-Based Polyester Depolymerization
Abstract
:1. Introduction
2. Results and Discussion
2.1. Production of the Recombinant PET2
2.2. PET2 Activity on Terephthalate Aromatic Polyesters
2.3. PET2 Activity on Indole-Based Polyesters
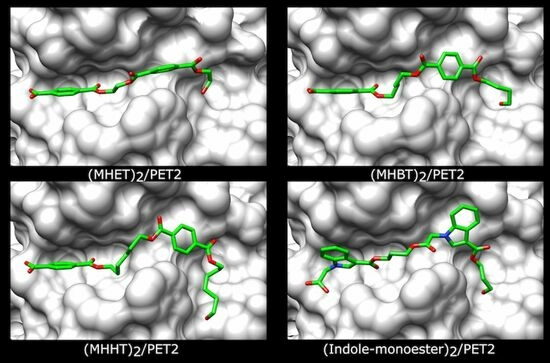
2.4. Structural Analysis
3. Materials and Methods
3.1. Production and Purification of Recombinant PET2 and Trx-IsPETase
3.2. MALDI-TOF/TOF
3.3. Polymer Preparation
3.4. Enzymatic Reactions
3.5. Reaction Products Analysis
3.6. Differential Scanning Fluorometry
3.7. Molecular Docking
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilcox, C.; Van Sebille, E.; Hardesty, B.D. Threat of plastic pollution to seabirds is global, pervasive, and increasing. Proc. Natl. Acad. Sci. USA 2015, 112, 11899–11904. [Google Scholar] [CrossRef]
- Kumar, R.; Sadeghi, K.; Jang, J.; Seo, J. Mechanical, chemical, and bio-recycling of biodegradable plastic: A review. Sci. Total Environ. 2023, 882, 163446. [Google Scholar] [CrossRef]
- Mankar, S.V.; Wahlberg, J.; Warlin, N.; Valsange, N.G.; Rehnberg, N.; Lundmark, S.; Jannasch, P.; Zhang, B. Short-Loop Chemical Recycling via Telechelic Polymers for Biobased Polyesters with Spiroacetal Units. ACS Sustain. Chem. Eng. 2023, 11, 5135–5146. [Google Scholar] [CrossRef]
- Arza, C.R.; Wang, P.; Linares-Pastén, J.; Zhang, B. Synthesis, thermal, rheological characteristics, and enzymatic degradation of aliphatic polyesters with lignin-based aromatic pendant groups. J. Polym. Sci. Part A Polym. Chem. 2019, 57, 2314–2323. [Google Scholar] [CrossRef]
- Nikiema, J.; Asiedu, Z. A review of the cost and effectiveness of solutions to address plastic pollution. Environ. Sci. Pollut. Res. 2022, 29, 24547–24573. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.J.; Schrader, H.; Profe, J.; Dresler, K.; Deckwer, W.D. Enzymatic degradation of poly (ethylene terephthalate): Rapid hydrolyse using a hydrolase from T. fusca. Macromol. Rapid Commun. 2005, 26, 1400–1405. [Google Scholar] [CrossRef]
- Kaabel, S.; Therien, J.D.; Deschênes, C.E.; Duncan, D.; Friščić, T.; Auclair, K. Enzymatic depolymerization of highly crystalline polyethylene terephthalate enabled in moist-solid reaction mixtures. Proc. Natl. Acad. Sci. USA 2021, 118, e2026452118. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.-L.; Texier, H.; Gavalda, S. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Danso, D.; Schmeisser, C.; Chow, J.; Zimmermann, W.; Wei, R.; Leggewie, C.; Li, X.; Hazen, T.; Streit, W.R. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl. Environ. Microbiol. 2018, 84, e02773-17. [Google Scholar] [CrossRef]
- Hufendiek, A.; Lingier, S.; Du Prez, F.E. Thermoplastic polyacetals: Chemistry from the past for a sustainable future? Polym. Chem. 2019, 10, 9–33. [Google Scholar] [CrossRef]
- Hatti-Kaul, R.; Nilsson, L.J.; Zhang, B.; Rehnberg, N.; Lundmark, S. Designing biobased recyclable polymers for plastics. Trends Biotechnol. 2020, 38, 50–67. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, F.; Lu, H.; Li, X.; Yang, X.; Tu, Y. A Cascade Polymerization Method for the Property Modification of Poly (butylene terephthalate) by the Incorporation of Isosorbide. ACS Appl. Polym. Mater. 2019, 1, 2313–2321. [Google Scholar] [CrossRef]
- De Vos, L.; Van de Voorde, B.; Van Daele, L.; Dubruel, P.; Van Vlierberghe, S. Poly (alkylene terephthalate) s: From current developments in synthetic strategies towards applications. Eur. Polym. J. 2021, 161, 110840. [Google Scholar] [CrossRef]
- Peng, B.-Y.; Zhang, X.; Sun, Y.; Liu, Y.; Chen, J.; Shen, Z.; Zhou, X.; Zhang, Y. Biodegradation and Carbon Resource Recovery of Poly (butylene adipate-co-terephthalate)(PBAT) by Mealworms: Removal Efficiency, Depolymerization Pattern, and Microplastic Residue. ACS Sustain. Chem. Eng. 2023, 11, 1774–1784. [Google Scholar] [CrossRef]
- Yang, Y.; Min, J.; Xue, T.; Jiang, P.; Liu, X.; Peng, R.; Huang, J.-W.; Qu, Y.; Li, X.; Ma, N. Complete bio-degradation of poly (butylene adipate-co-terephthalate) via engineered cutinases. Nat. Commun. 2023, 14, 1645. [Google Scholar] [CrossRef]
- Guo, Z.; Warlin, N.; Mankar, S.V.; Sidqi, M.; Andersson, M.; Zhang, B.; Nilsson, E. Development of Circularly Recyclable Low Melting Temperature Bicomponent Fibers toward a Sustainable Nonwoven Application. ACS Sustain. Chem. Eng. 2021, 9, 16778–16785. [Google Scholar] [CrossRef]
- Qian, K.; Qiao, R.; Chen, S.; Luo, H.; Zhang, D. Enhanced permittivity in polymer blends via tailoring the orderliness of semiconductive liquid crystalline polymers and intermolecular interactions. J. Mater. Chem. C 2020, 8, 8440–8450. [Google Scholar] [CrossRef]
- Hall, I.; Ibrahim, B. The structure and properties of poly (hexamethylene terephthalate): 1. The preparation, morphology and unit cells of three allomorphs. Polymer 1982, 23, 805–816. [Google Scholar] [CrossRef]
- Pan, T.; Deng, J.; Xu, Q.; Zuo, Y.; Guo, Q.X.; Fu, Y. Catalytic Conversion of Furfural into a 2, 5-Furandicarboxylic Acid-Based Polyester with Total Carbon Utilization. ChemSusChem 2013, 6, 47–50. [Google Scholar] [CrossRef]
- Amarasekara, A.S.; Nguyen, L.H.; Okorie, N.C.; Jamal, S.M. A two-step efficient preparation of a renewable dicarboxylic acid monomer 5, 5′-[oxybis (methylene)] bis [2-furancarboxylic acid] from d-fructose and its application in polyester synthesis. Green Chem. 2017, 19, 1570–1575. [Google Scholar] [CrossRef]
- Rosenboom, J.-G.; Hohl, D.K.; Fleckenstein, P.; Storti, G.; Morbidelli, M. Bottle-grade polyethylene furanoate from ring-opening polymerisation of cyclic oligomers. Nat. Commun. 2018, 9, 2701. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Xu, Y.; Jiang, M.; Wang, R.; Wang, H.; Liang, Y.; Zhou, G. Fully bio-based polyesters poly (ethylene-co-1, 5-pentylene 2, 5-thiophenedicarboxylate) s (PEPTs) with high toughness: Synthesis, characterization and thermo-mechanical properties. Polymer 2020, 204, 122800. [Google Scholar] [CrossRef]
- Nguyen, H.T.H.; Reis, M.H.; Qi, P.; Miller, S.A. Polyethylene ferulate (PEF) and congeners: Polystyrene mimics derived from biorenewable aromatics. Green Chem. 2015, 17, 4512–4517. [Google Scholar] [CrossRef]
- Gioia, C.; Banella, M.B.; Marchese, P.; Vannini, M.; Colonna, M.; Celli, A. Advances in the synthesis of bio-based aromatic polyesters: Novel copolymers derived from vanillic acid and ε-caprolactone. Polym. Chem. 2016, 7, 5396–5406. [Google Scholar] [CrossRef]
- Wurdemann, M.A.; Bernaerts, K.V. Biobased Pyrazine-Containing Polyesters. ACS Sustain. Chem. Eng. 2020, 8, 12045–12052. [Google Scholar] [CrossRef]
- Burgess, S.K.; Karvan, O.; Johnson, J.; Kriegel, R.M.; Koros, W.J. Oxygen sorption and transport in amorphous poly (ethylene furanoate). Polymer 2014, 55, 4748–4756. [Google Scholar] [CrossRef]
- Terzopoulou, Z.; Karakatsianopoulou, E.; Kasmi, N.; Tsanaktsis, V.; Nikolaidis, N.; Kostoglou, M.; Papageorgiou, G.Z.; Lambropoulou, D.A.; Bikiaris, D.N. Effect of catalyst type on molecular weight increase and coloration of poly (ethylene furanoate) biobased polyester during melt polycondensation. Polym. Chem. 2017, 8, 6895–6908. [Google Scholar] [CrossRef]
- Loos, K.; Zhang, R.; Pereira, I.; Agostinho, B.; Hu, H.; Maniar, D.; Sbirrazzuoli, N.; Silvestre, A.; Guigo, N.; Sousa, A. A Perspective on PEF Synthesis, Properties, and End-Life. Front. Chem. 2020, 8, 585. [Google Scholar] [CrossRef]
- Tsanaktsis, V.; Terzopoulou, Z.; Exarhopoulos, S.; Bikiaris, D.N.; Achilias, D.S.; Papageorgiou, D.G.; Papageorgiou, G.Z. Sustainable, eco-friendly polyesters synthesized from renewable resources: Preparation and thermal characteristics of poly (dimethyl-propylene furanoate). Polym. Chem. 2015, 6, 8284–8296. [Google Scholar] [CrossRef]
- Ma, Q.; Zhang, X.; Qu, Y. Biodegradation and biotransformation of indole: Advances and perspectives. Front. Microbiol. 2018, 9, 2625. [Google Scholar] [CrossRef]
- Wang, P.; Arza, C.R.; Zhang, B. Indole as a new sustainable aromatic unit for high quality biopolyesters. Polym. Chem. 2018, 9, 4706–4710. [Google Scholar] [CrossRef]
- Wang, P.; Linares-Pastén, J.A.; Zhang, B. Synthesis, Molecular Docking Simulation, and Enzymatic Degradation of AB-Type Indole-Based Polyesters with Improved Thermal Properties. Biomacromolecules 2020, 21, 1078–1090. [Google Scholar] [CrossRef] [PubMed]
- Arza, C.R.; Zhang, B. Synthesis, thermal properties, and rheological characteristics of indole-based aromatic polyesters. ACS Omega 2019, 4, 15012–15021. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, B. Sustainable aromatic polyesters with 1, 5-disubstituted indole units. RSC Adv. 2021, 11, 16480–16489. [Google Scholar] [CrossRef]
- Wagner-Egea, P.; Tosi, V.; Wang, P.; Grey, C.; Zhang, B.; Linares-Pastén, J.A. Assessment of IsPETase-Assisted Depolymerization of Terephthalate Aromatic Polyesters and the Effect of the Thioredoxin Fusion Domain. Appl. Sci. 2021, 11, 8315. [Google Scholar] [CrossRef]
- Meilleur, C.; Hupé, J.-F.; Juteau, P.; Shareck, F. Isolation and characterization of a new alkali-thermostable lipase cloned from a metagenomic library. J. Ind. Microbiol. Biotechnol. 2009, 36, 853–861. [Google Scholar] [CrossRef]
- Nakamura, A.; Kobayashi, N.; Koga, N.; Iino, R. Positive charge introduction on the surface of thermostabilized PET hydrolase facilitates PET binding and degradation. ACS Catal. 2021, 11, 8550–8564. [Google Scholar] [CrossRef]
- Aristizábal-Lanza, L.; Mankar, S.V.; Tullberg, C.; Zhang, B.; Linares-Pastén, J.A. Comparison of the enzymatic depolymerization of polyethylene terephthalate and AkestraTM using Humicola insolens cutinase. Front. Chem. Eng. 2022, 4, 1048744. [Google Scholar] [CrossRef]
- Joo, S.; Cho, I.J.; Seo, H.; Son, H.F.; Sagong, H.-Y.; Shin, T.J.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Structural insight into molecular mechanism of poly (ethylene terephthalate) degradation. Nat. Commun. 2018, 9, 382. [Google Scholar] [CrossRef]
- Boneta, S.; Arafet, K.; Moliner, V. QM/MM Study of the Enzymatic Biodegradation Mechanism of Polyethylene Terephthalate. J. Chem. Inf. Model. 2021, 61, 3041–3051. [Google Scholar] [CrossRef]
- Jerves, C.; Neves, R.P.; Ramos, M.J.; da Silva, S.; Fernandes, P.A. Reaction mechanism of the PET degrading enzyme PETase studied with DFT/MM molecular dynamics simulations. ACS Catal. 2021, 11, 11626–11638. [Google Scholar] [CrossRef]
- Richter, P.K.; Blázquez-Sánchez, P.; Zhao, Z.; Engelberger, F.; Wiebeler, C.; Künze, G.; Frank, R.; Krinke, D.; Frezzotti, E.; Lihanova, Y. Structure and function of the metagenomic plastic-degrading polyester hydrolase PHL7 bound to its product. Nat. Commun. 2023, 14, 1905. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Song, C.; Gräsing, D.; Schneider, T.; Bielytskyi, P.; Böttcher, D.; Matysik, J.; Bornscheuer, U.T.; Zimmermann, W. Conformational fitting of a flexible oligomeric substrate does not explain the enzymatic PET degradation. Nat. Commun. 2019, 10, 5581. [Google Scholar] [CrossRef] [PubMed]
- Sagong, H.-Y.; Seo, H.; Kim, T.; Son, H.F.; Joo, S.; Lee, S.H.; Kim, S.; Woo, J.-S.; Hwang, S.Y.; Kim, K.-J. Decomposition of the PET film by MHETase using Exo-PETase function. ACS Catal. 2020, 10, 4805–4812. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Warlin, N.; Gonzalez, M.N.G.; Mankar, S.; Valsange, N.G.; Sayed, M.; Pyo, S.-H.; Rehnberg, N.; Lundmark, S.; Hatti-Kaul, R.; Jannasch, P. A rigid spirocyclic diol from fructose-based 5-hydroxymethylfurfural: Synthesis, life-cycle assessment, and polymerization for renewable polyesters and poly (urethane-urea) s. Green Chem. 2019, 21, 6667–6684. [Google Scholar] [CrossRef]
- Din, S.U.; Satti, S.M.; Uddin, S.; Mankar, S.V.; Ceylan, E.; Hasan, F.; Khan, S.; Badshah, M.; Beldüz, A.O.; Çanakçi, S.; et al. The Purification and Characterization of a Cutinase-like Enzyme with Activity on Polyethylene Terephthalate (PET) from a Newly Isolated Bacterium Stenotrophomonas maltophilia PRS8 at a Mesophilic Temperature. Appl. Sci. 2023, 13, 3686. [Google Scholar] [CrossRef]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins Struct. Funct. Bioinform. 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Li, X.; Ilk, S.; Linares-Pastén, J.A.; Liu, Y.; Raina, D.B.; Demircan, D.; Zhang, B. Synthesis, Enzymatic Degradation, and Polymer-Miscibility Evaluation of Nonionic Antimicrobial Hyperbranched Polyesters with Indole or Isatin Functionalities. Biomacromolecules 2021, 22, 2256–2271. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]








| Polymer | PET2 | IsPETase [35] | ||
|---|---|---|---|---|
| Degradation Products (mg/L) | Depolymerization (%) | Degradation Products (mg/L) | Depolymerization (%) | |
| PET | 1965 | 19.66 | 3934 | 39.34 |
| PBT | 138 | 1.38 | 25 | 0.25 |
| PHT | 974 | 9.74 | 12.5 | 0.13 |
| Akestra™ | N.D. | N.D. | 13.3 | 0.13 |
| AB | N.D. | N.D. | ||
| AABB | 925 | 7.01 | N.Q. | N.Q. |
| Polymer | IV * (dL/g) | Tg (°C) | Tm (°C) | Mn (g/mol) | Mw (g/mol) | Crystallinity (%) | Reference |
|---|---|---|---|---|---|---|---|
| PET | ≈0.8 | ≈78 | ≈245 | 11.3 | [35] | ||
| PBT | ≈203 | ≈38,000 | 38.6 | [35] | |||
| PHT | ≈1.26 | ≈17 | ≈140 | ≈26,000 | ≈43,000 | n.d. | [35] |
| Akestra™ | ≈0.64 | ≈95 | - | Amorphous | [35] | ||
| AA | 102 | - | ≈8100 | ≈14,500 | Amorphous | [32] | |
| AABB | 93 | - | ≈35,800 | ≈71,700 | Amorphous | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagner-Egea, P.; Aristizábal-Lanza, L.; Tullberg, C.; Wang, P.; Bernfur, K.; Grey, C.; Zhang, B.; Linares-Pastén, J.A. Marine PET Hydrolase (PET2): Assessment of Terephthalate- and Indole-Based Polyester Depolymerization. Catalysts 2023, 13, 1234. https://doi.org/10.3390/catal13091234
Wagner-Egea P, Aristizábal-Lanza L, Tullberg C, Wang P, Bernfur K, Grey C, Zhang B, Linares-Pastén JA. Marine PET Hydrolase (PET2): Assessment of Terephthalate- and Indole-Based Polyester Depolymerization. Catalysts. 2023; 13(9):1234. https://doi.org/10.3390/catal13091234
Chicago/Turabian StyleWagner-Egea, Paula, Lucía Aristizábal-Lanza, Cecilia Tullberg, Ping Wang, Katja Bernfur, Carl Grey, Baozhong Zhang, and Javier A. Linares-Pastén. 2023. "Marine PET Hydrolase (PET2): Assessment of Terephthalate- and Indole-Based Polyester Depolymerization" Catalysts 13, no. 9: 1234. https://doi.org/10.3390/catal13091234