CalkGH9T: A Glycoside Hydrolase Family 9 Enzyme from Clostridium alkalicellulosi
Abstract
:1. Introduction
2. Results and Discussion
2.1. Bioinformatics Analysis
2.2. Protein Expression and Purification
2.3. Substrate Preference and Effects of pH and Temperature on Enzyme Activity
2.4. Effect of Metal Ions and Chemicals
2.5. Effect of NaCl Concentration
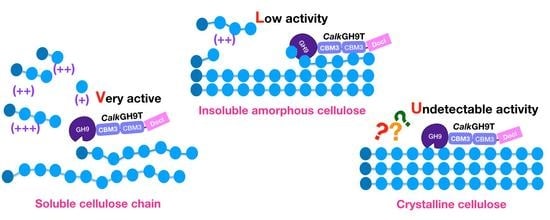
2.6. Mode of Action
2.6.1. Activity on Polysaccharides
2.6.2. Activity on Cellodextrins
2.7. Cohesin-Dockerin Interaction
2.8. Binding Ability to Polysaccharides
2.9. Processive Activity
2.10. Addition of CalkGH9T to a Fungal Enzyme Preparation
3. Materials and Methods
3.1. DNA, Bacterial Strains, Plasmid, and Chemicals
3.2. Gene Cloning and Recombinant DNA Techniques
3.3. Protein Expression and Purification
3.4. Enzyme Activity Assay
3.5. Effect of pH and Temperature
3.6. Effect of Metal Ions, Salts, and Chemicals
3.7. Analysis of the Hydrolysis Product
3.8. Cohesin-Dockerin Interaction Test Using Enzyme-Linked Immunosorbent Assay (ELISA)
3.9. Polysaccharide Binding Assay
3.10. Processive Activity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hamid, S.B.A.; Islam, M.M.; Das, R. Cellulase biocatalysis: Key influencing factors and mode of action. Cellulose 2015, 22, 2157–2182. [Google Scholar] [CrossRef]
- Phitsuwan, P.; Sakka, K.; Ratanakhanokchai, K. Improvement of lignocellulosic biomass in planta:a review of feedstocks, biomass recalcitrance, and strategic manipulation of ideal plants designed for ethanol production and processability. Biomass Bioenergy 2013, 58, 390–405. [Google Scholar] [CrossRef]
- Gupta, V.K.; Steindorff, A.S.; de Paula, R.G.; Silva-Rocha, R.; Mach-Aigner, A.R.; Mach, R.L.; Silva, R.N. The post-genomic era of Trichoderma reesei: What’s next? Trends Biotechnol. 2016, 34, 970–982. [Google Scholar] [CrossRef] [Green Version]
- Akram, F.; Haq, I.u.; Imran, W.; Mukhtar, H. Insight perspectives of thermostable endoglucanases for bioethanol production: A review. Renew. Energy 2018, 122, 225–238. [Google Scholar] [CrossRef]
- Beckham, G.T.; Ståhlberg, J.; Knott, B.C.; Himmel, M.E.; Crowley, M.F.; Sandgren, M.; Sørlie, M.; Payne, C.M. Towards a molecular-level theory of carbohydrate processivity in glycoside hydrolases. Curr. Opin. Biotechnol. 2014, 27, 96–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravachol, J.; Borne, R.; Tardif, C.; De Philip, P.; Fierobe, H.P. Characterization of all family-9 glycoside hydrolases synthesized by the cellulosome-producing bacterium Clostridium cellulolyticum. J. Biol. Chem. 2014, 289, 7335–7348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arai, T.; Kosugi, A.; Chan, H.; Koukiekolo, R.; Yukawa, H.; Inui, M.; Doi, R.H. Properties of cellulosomal family 9 cellulases from Clostridium cellulovorans. Appl. Microbiol. Biotechnol. 2006, 71, 654–660. [Google Scholar] [CrossRef]
- Petkun, S.; Jindou, S.; Shimon, L.J.W.; Rosenheck, S.; Bayer, E.A.; Lamed, R.; Frolow, F. Structure of a family 3b’ carbohydrate-binding module from the Cel9V glycoside hydrolase from Clostridium thermocellum: Structural diversity and implications for carbohydrate binding. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 33–43. [Google Scholar] [CrossRef]
- Schröder, C.; Burkhardt, C.; Busch, P.; Schirrmacher, G.; Claren, J.; Antranikian, G. Characterization of a theme C glycoside hydrolase family 9 endo-β-Glucanase from a biogas reactor metagenome. Protein J. 2018, 37, 454–460. [Google Scholar] [CrossRef]
- Zhang, K.D.; Li, W.; Wang, Y.F.; Zheng, Y.L.; Tan, F.C.; Ma, X.Q.; Yao, L.S.; Bayer, E.A.; Wang, L.S.; Li, F.L. Processive degradation of crystalline cellulose by a multimodular endoglucanase via a wirewalking mode. Biomacromolecules 2018, 19, 1686–1696. [Google Scholar] [CrossRef]
- Mingardon, F.; Bagert, J.D.; Maisonnier, C.; Trudeau, D.L.; Arnold, F.H. Comparison of family 9 cellulases from mesophilic and thermophilic bacteria. Appl. Environ. Microbiol. 2011, 77, 1436–1442. [Google Scholar] [CrossRef] [Green Version]
- Qi, M.; Jun, H.S.; Forsberg, C.W. Cel9D, an atypical 1,4-β-D-glucan glucohydrolase from Fibrobacter succinogenes: Characteristics, catalytic residues, and synergistic interactions with other cellulases. J. Bacteriol. 2008, 190, 1976–1984. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Irwin, D.C.; Wilson, D.B. Processivity, substrate binding, and mechanism of cellulose hydrolysis by Thermobifida fusca Cel9A. Appl. Environ. Microbiol. 2007, 73, 3165–3172. [Google Scholar] [CrossRef] [Green Version]
- Gilad, R.; Rabinovich, L.; Yaron, S.; Bayer, E.A.; Lamed, R.; Gilbert, H.J.; Shoham, Y. Cell, a noncellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J. Bacteriol. 2003, 185, 391–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeng, W.Y.; Liu, C.I.; Lu, T.J.; Lin, H.J.; Wang, N.C.; Wang, A.H.J. Crystal structures of the C-terminally truncated endoglucanase Cel9Q from Clostridium thermocellum complexed with cellodextrins and Tris. ChemBioChem 2019, 20, 295–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jindou, S.; Xu, Q.; Kenig, R.; Shulman, M.; Shoham, Y.; Bayer, E.A.; Lamed, R. Novel architecture of family-9 glycoside hydrolases identified in cellulosomal enzymes of Acetivibrio cellulolyticus and Clostridium thermocellum. FEMS Microbiol. Lett. 2006, 254, 308–316. [Google Scholar] [CrossRef] [Green Version]
- Artzi, L.; Morag, E.; Barak, Y.; Lamed, R.; Bayer, E.A. Clostridium clariflavum: Key cellulosome players are revealed by proteomic analysis. mBio 2015, 6, e00411–e00415. [Google Scholar] [CrossRef] [Green Version]
- Zvereva, E.A.; Fedorova, T.V.; Kevbrin, V.V.; Zhilina, T.N.; Rabinovich, M.L. Cellulase activity of a haloalkaliphilic anaerobic bacterium, strain Z-7026. Extremophiles 2006, 10, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Zhilina, T.N.; Kevbrin, V.V.; Tourova, T.P.; Lysenko, A.M.; Kostrikina, N.A.; Zavarzin, G.A. Clostridium alkalicellum sp. nov., an obligately alkaliphilic cellulolytic bacterium from a soda lake in the Baikal region. Microbiology 2005, 74, 557–566. [Google Scholar] [CrossRef]
- Artzi, L.; Bayer, E.A.; Moraïs, S. Cellulosomes: Bacterial nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 2017, 15, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Phitsuwan, P.; Moraïs, S.; Dassa, B.; Henrissat, B.; Bayer, E.A. The cellulosome paradigm in an extreme alkaline environment. Microorganisms 2019, 7, 347. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurokawa, J.; Hemjinda, E.; Arai, T.; Kimura, T.; Sakka, K.; Ohmiya, K. Clostridium thermocellum cellulase CelT, a family 9 endoglucanase without an Ig-like domain or family 3c carbohydrate-binding module. Appl. Microbiol. Biotechnol. 2002, 59, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Petkun, S.; Rozman Grinberg, I.; Lamed, R.; Jindou, S.; Burstein, T.; Yaniv, O.; Shoham, Y.; Shimon, L.J.; Bayer, E.A.; Frolow, F. Reassembly and co-crystallization of a family 9 processive endoglucanase from its component parts: Structural and functional significance of the intermodular linker. PeerJ 2015, 3, e1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.J.; Kim, S.H.; Shin, S.K.; Hyeon, J.E.; Han, S.O. Mutation of a conserved tryptophan residue in the CBM3c of a GH9 endoglucanase inhibits activity. Int. J. Biol. Macromol. 2016, 92, 159–166. [Google Scholar] [CrossRef]
- Montanier, C.; Money, V.A.; Pires, V.M.; Flint, J.E.; Pinheiro, B.A.; Goyal, A.; Prates, J.A.; Izumi, A.; Stalbrand, H.; Morland, C.; et al. The active site of a carbohydrate esterase displays divergent catalytic and noncatalytic binding functions. PLoS Biol. 2009, 7, e71. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Ruan, L.; Chen, X.; Zhang, Y.; Xu, X. A novel salt-tolerant endo-β-1,4-glucanase Cel5A in Vibrio sp. G21 isolated from mangrove soil. Appl. Microbiol. Biotechnol. 2010, 87, 1373–1382. [Google Scholar] [CrossRef]
- Percival Zhang, Y.H.; Himmel, M.E.; Mielenz, J.R. Outlook for cellulase improvement: Screening and selection strategies. Biotechnol. Adv. 2006, 24, 452–481. [Google Scholar] [CrossRef]
- Eckert, K.; Ernst, H.A.; Schneider, E.; Larsen, S.; Lo Leggio, L. Crystallization and preliminary X-ray analysis of Alicyclobacillus acidocaldarius endoglucanase CelA. Acta Crystallogr. D Biol. Crystallogr. 2003, 59, 139–141. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.Y.; Li, X. Alkali-stable cellulase from a halophilic isolate, Gracilibacillus sp. SK1 and its application in lignocellulosic saccharification for ethanol production. Biomass Bioenergy 2015, 81, 19–25. [Google Scholar] [CrossRef]
- Xue, D.S.; Liang, L.Y.; Lin, D.Q.; Gong, C.J.; Yao, S.J. Halostable catalytic properties of exoglucanase from a marine Aspergillus niger and secondary structure change caused by high salinities. Process Biochem. 2017, 58, 85–91. [Google Scholar] [CrossRef]
- Danson, M.J.; Hough, D.W. The structural basis of protein halophilicity. Comp. Biochem. Physiol. A Physiol. 1997, 117, 307–312. [Google Scholar] [CrossRef]
- Artzi, L.; Dassa, B.; Borovok, I.; Shamshoum, M.; Lamed, R.; Bayer, E.A. Cellulosomics of the cellulolytic thermophile Clostridium clariflavum. Biotechnol. Biofuels 2014, 7, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamberg, Y.; Ruimy-Israeli, V.; Dassa, B.; Barak, Y.; Lamed, R.; Cameron, K.; Fontes, C.M.; Bayer, E.A.; Fried, D.B. Elaborate cellulosome architecture of Acetivibrio cellulolyticus revealed by selective screening of cohesin-dockerin interactions. PeerJ 2014, 2, e636. [Google Scholar] [CrossRef]
- Zhang, X.Z.; Sathitsuksanoh, N.; Zhang, Y.H.P. Glycoside hydrolase family 9 processive endoglucanase from Clostridium phytofermentans: Heterologous expression, characterization, and synergy with family 48 cellobiohydrolase. Bioresour. Technol. 2010, 101, 5534–5538. [Google Scholar] [CrossRef] [PubMed]
- Irwin, D.C.; Spezio, M.; Walker, L.P.; Wilson, D.B. Activity studies of eight purified cellulases: Specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 1993, 42, 1002–1013. [Google Scholar] [CrossRef]
- Kruus, K.; Wang, W.K.; Ching, J.; Wu, J.H.D. Exoglucanase activities of the recombinant Clostridium thermocellum CelS, a major cellulosome component. J. Bacteriol. 1995, 177, 1641–1644. [Google Scholar] [CrossRef] [Green Version]
- Payne, C.M.; Knott, B.C.; Mayes, H.B.; Hansson, H.; Himmel, M.E.; Sandgren, M.; Ståhlberg, J.; Beckham, G.T. Fungal cellulases. Chem. Rev. 2015, 115, 1308–1448. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.H.P.; Cui, J.; Lynd, L.R.; Kuang, L.R. A Transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules 2006, 7, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Barak, Y.; Handelsman, T.; Nakar, D.; Mechaly, A.; Lamed, R.; Shoham, Y.; Bayer, E.A. Matching fusion protein systems for affinity analysis of two interacting families of proteins: The cohesin-dockerin interaction. J. Mol. Recognit. 2005, 18, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]









| Substrate | Main Linkage | Solubility in Water | Activity (µM Product/min/µM Protein) | Relative Activity (%) |
|---|---|---|---|---|
| CMC | β-1,4 glucan | Soluble | 1021 | 100 |
| RAC | β-1,4 glucan | Insoluble | 142 1 | 14 |
| Avicel | β-1,4 glucan | Insoluble | ND 2 | ND |
| Xylan | β-1,4 xylan | In/soluble | 261 | 26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phitsuwan, P.; Lee, S.; San, T.; Ratanakhanokchai, K. CalkGH9T: A Glycoside Hydrolase Family 9 Enzyme from Clostridium alkalicellulosi. Catalysts 2021, 11, 1011. https://doi.org/10.3390/catal11081011
Phitsuwan P, Lee S, San T, Ratanakhanokchai K. CalkGH9T: A Glycoside Hydrolase Family 9 Enzyme from Clostridium alkalicellulosi. Catalysts. 2021; 11(8):1011. https://doi.org/10.3390/catal11081011
Chicago/Turabian StylePhitsuwan, Paripok, Sengthong Lee, Techly San, and Khanok Ratanakhanokchai. 2021. "CalkGH9T: A Glycoside Hydrolase Family 9 Enzyme from Clostridium alkalicellulosi" Catalysts 11, no. 8: 1011. https://doi.org/10.3390/catal11081011