1. Introduction
Methanotrophs can be used for various purposes, such as purification of environments polluted by chlorinated hydrocarbons or harmful heavy metals [
1,
2,
3], removal of methane, which is a more potent greenhouse gas than carbon dioxide [
4], biological nitrogen fixation and denitrification [
5], production of bio-methanol, [
6,
7,
8,
9,
10,
11,
12,
13,
14,
15], and production of bioplastics, such as poly-3-hydroxybutyrate (PHB) [
16,
17]. Methanotrophs are bacteria that play an important role in the sustainability of the global environment. Previous studies on methanol production by methanotrophs mainly focused on a single species (mainly type II methanotrophs) and used nitrate mineral salts medium (NMS medium) or ammonium mineral salts medium (AMS medium) [
18], which are chemically defined culture media [
6,
7,
8,
9,
10,
11,
12,
13,
14,
15]. However, chemically defined culture media and commercial media are expensive and generate chemical wastewater. In addition, to create the optimum conditions for a specific single species to enable it to continuously survive as an absolutely dominant species, manipulations, such as sterilization and blocking of external contaminations, are necessary, which are difficult to be carried out in uncontrolled environments outside the laboratory.
To date, high-purity methane (CH
4 > 97%) has been used in the production of methanol by methanotrophs. Such production requires purification and separation processes, which are costly. In other words, previous studies lack consideration of realistic alternatives for economic feasibility and can only be conducted in controlled laboratory settings. To improve economic feasibility, certain studies have used biogas, instead of high-purity methane, as a realistic alternative, while others have sought to produce methanol from methane, among other gases that form crude biogas [
15,
16,
17,
18,
19,
20]. However, these studies also targeted specific species to methanotrophs, limiting the application of the methods outside of laboratories.
In a previous study, for the first time, we used sewage flowing into an urban wastewater treatment plant as a culture medium, as well as biogas generated from a wastewater treatment plant in bio-methanol production [
21]. In subsequent studies, we produced methanol using biogas as a carbon source and sewage as a culture medium and proposed a treatment process using the generated methanol as a carbon source for denitrification [
22,
23]. Furthermore, we investigated the bacterial community formation and the dominance of effectives species in reject water containing high concentrations of ammonia generated by anaerobic digesters in wastewater treatment plants, as well as the mutual roles and correlations between each bacterial cluster involved in the process [
24].
Compared to previous studies that used methane (a pure synthetic gas) and chemically defined or commercially sold culture media, our previous studies using biogas generated by wastewater treatment plants as a carbon source, as well as nutrient-rich sewage and wastewater as culture media, were different [
21,
22,
23,
24]. The biggest difference was that maintaining an effective single species as a dominant species, using wastewater and biogas, was not possible owing to biological contamination and interference, unlike in controlled laboratory environments. Bacterial clusters of different taxa accumulated to form a bacterial community in which the bacteria are interdependent. The formed bacterial communities were useful in methanol production and wastewater processing [
21,
22,
23,
24]. Another key difference from controlled laboratory conditions, where only pure methane was used, was the existence of new effectives species under high CO
2 ratio of ≥30%. The key species in the bacterial community were the
Methylophilus strains that used the methanol produced by methanotrophs. Additionally, new species of methanotrophs classified as type X have been observed in experiments using biogas, compared with those using only methane [
24]. We observed that type X methanotrophs could convert CO
2 into methanol if a reducing equivalent was present in the reaction system [
25,
26,
27,
28]. In the current study, for the effective use of CO
2 in biological carbon fixation and wastewater treatment and for carbon capture, utilization, and storage (CCUS), we explored biological carbon fixation using bacterial community and biogas.
The concept of CO
2 fixation by methanotrophs first emerged following the discovery a new methanotroph species that produces methanol from CO
2 through the Calvin–Benson–Bassham (CBB) cycle in the Verrumicrobia phylum. However, this species is a thermophilic methanotroph that inhabits high-temperature environments, such as volcanoes and hot springs; thus, it cannot be widely used for CO
2 fixation [
25,
26,
27]. However, some methanotrophs, such as type X methanotrophs, may be induced to become parts of bacterial community and use CO
2 as a substrate to reduce excess CO
2 into methanol [
28]. In the first step of the process, methanotrophs proliferate using methane as a carbon substrate. In the second step, CO
2 is reduced to methanol by the generated methanotrophs. Reduction of CO
2 to methanol is the reverse reaction of methane oxidation to CO
2. To reduce CO
2 to methanol, a high reducing power is required to induce a reaction that is against the law of energy. In other words, NADH is required to reduce CO
2 to formaldehyde, and pyrroloquinoline quinol (PQQH2) is needed to reduce formaldehyde to methanol [
25,
26,
27,
28].
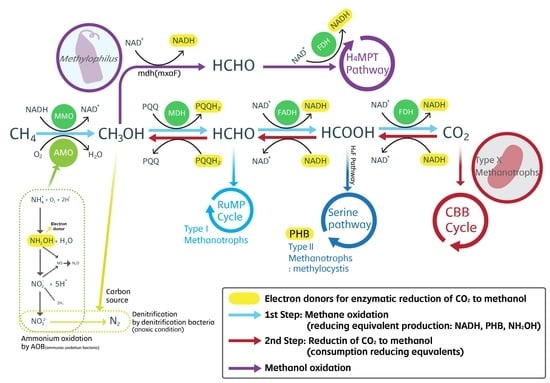
In this study, we aimed to assess the optimum conditions for generating the driving force of reverse reaction against the law of energy in bacterial community through experiments, as well as to use this reaction to biologically fix and remove greenhouse gases, such as CH4 and CO2, for effective use in wastewater treatment. For these purposes, we efficiently maintained the bacterial community using biogas mainly consisting of CH4 and CO2 instead of manufactured or purified CH4 as carbon sources for bacteria. We also established the optimum conditions for generating methanotrophs capable of using CO2 as a carbon source, such as type X methanotrophs, as the dominant species in the bacterial community while efficiently and continuously providing the reducing power required to convert CO2 to methanol.
2. Results and Discussion
2.1. Characteristics of Cultured Microbial Community
As described in
Section 3.1, activated sludge from wastewater treatment plants was cultured in AMS medium with biogas obtained from a wastewater treatment plant, in a pressure vessel to incubate bacteria for subsequent experiments. CH
4, CO
2, and O
2 were mixed at a ratio of 1:0.5:1 in AMS medium. For each 24 h cycle, fresh AMS medium and fresh mix of the gases were provided to culture the bacterial communities. The results are shown in
Figure 1.
Figure 1A shows the proportion and abundance percentage of each strain of the bacterial community at the class and genus levels.
Figure 1B shows an electron microscopy image of the bacterial community.
Bacillus and
Micrococcus, which are typical type I methanotrophs, showed a high abundance [
29].
Among the observed methanotroph strains,
Methylobacter showed a high abundance, accounting for approximately 42.3% of the total bacterial community. The number of
Methylophilus strains, whose growth is dependent on the methanol produced by methanotrophs, was also high at 42.9%. These findings suggested
Methylobacter and
Methylophilus as the dominant species of the bacterial community. To assess differences in methanotroph strains when using biogas and pure methane, we compared the composition of methanotroph strains in the bacterial communities following culture with CH
4 and with biogas containing a high CO
2 content of ≥30% (31.3%). The results are presented in
Figure 2.
In both conditions, Methylobacter was the most dominant strain. However, the most significant difference between the two conditions was that type X methanotrophs were detected significantly at 11.2% only in the community cultured with CO2-containing biogas.
Figure 3 shows the distribution of strains according to a phylogenetic diagram of each bacterial cluster (type I, type II, and type X methanotrophs; methanol-oxidizing bacteria, with
Methylophilus as the dominant strain; and ammonia-oxidizing bacteria) in the bacterial community cultured with biogas. Type I and X methanotrophs, which are γ-proteobacteria, and type II methanotrophs, which are α-protobacteria, were classified at the genus level.
Methylophilus, which are closely dependent on methanotrophs [
30], are also shown in the
Figure 3. They are ammonia-oxidizing bacteria that partially oxidize methane through ammonia monooxygenase [
31], and they use ammonia as an energy source.
Table 1 categorizes the type X methanotroph strains as γ-proteobacteria and α-protobacteria.
Type I and II methanotrophs, as well as type X methanotrophs (Methylococcacea, Methylococcales), which belong to the γ-proteobacteria group, were identified as the target species that can convert CO
2 to methanol through reverse reduction. Additionally, the α-protobacteria
Methylocystis, were the only type II methanotrophs identified. Unlike type I methanotrophs, which express particulate methane monooxygenase (pMMO), type II methanotrophs express soluble methane monooxygenase (sMMO) and are not activated in the presence of copper [
32]. In this study, copper (as CuSO
4) concentration in the AMS medium was 20 μM, which is relatively high for the activation of type II methanotrophs, and extraordinarily, the type II methanotrophs
Methylocystis were activated.
A previous study by Muhammad and colleagues [
33] may explain why
Methylocystis strain was activated in our study. sMMO expression in
Methylocystis sp. strain SB2 is not greatly affected by copper concentration, unlike that in other type II methanotrophs. In that study, the expression of methanobactins (mbnA) in
Methylosinus trichosporium OB3b was not affected by copper concentration; regardless of copper concentration (1 μM CuCl
2) in the culture, sMMO was expressed to encode a precursor polypeptide of methanobactin. Moreover,
Methylocystis also have a higher capacity to synthesize poly-3-hydroxybutyrate (PHB) than other type II methanotrophs [
34]. In addition to NADH, PHB also acts as an electron donor for the reduction in CO
2, as it is a source of intracellular reducing equivalents. The NAD
+-linked enzyme β-hydroxybutyrate dehydrogenase catalyzes PHB degradation into acetoacetic acid, which consequently results in reducing equivalents. Therefore, the PHB stored in the type II methanotroph
Methylocystis plays an important role in the reduction of CO
2 into methanol [
34]. Thus, the coexistence of
Methylocystis with type X methanotrophs under the culture conditions of this study is of great importance.
In particular, Bashir and colleagues [
35] treated
Methylocystis strains with anaerobic digester-derived biogas or waste landfill gas for biological synthesis of PHB, which is an important raw material for biodegradable plastics. The PHB yield was up to 55.7 ± 1.9% of the dry weight of the bacterial cells. Two new
Methylocystis strains with high efficacy were observed:
Methylocystis rosea BRCS1 and
Methylocystis parvus BRCS2. As shown in
Figure 4, these are the same species observed in the bacterial community cultured with biogas in our study. Thus, we identified bacterial strains capable of providing reducing equivalents to reduce CO
2 through the generation of PHB.
2.2. Changes in Methanotrophs Strains Composition According to CO2 Increase
Section 2.1 describes the results of experiments using biogas from a wastewater treatment plant (CH
4: 65.2%, CO
2: 33.1%, N
2: 1.6%, O
2: 0.1%). The biogas was placed in a 350 mL flask containing 200 mL of AMS medium, and then the biogas and air content were adjusted in the extra 150 mL head space using a mixture gas containing CH
4, CO
2, and O
2 at a ratio of 1:0.5:1. In this section, the CH
4/O
2 ratio was fixed at 1:1, and the CO
2/CH
4 ratio was changed to 0.5, 1.0, 1.5, 2.0, and 2.5 to assess changes in bacteria strains, metabolites (formaldehyde, formate), and coenzyme ratio (NADH/NAD
+).
As the CO
2/CH
4 ratio gradually increased to 0.5, 1.0, 1.5, 2.0, and 2.5, the proportion of the type X methanotrophs
Methylococcus and
Methylocaldum in the bacterial community at the final 96 h of culture increased to 3.2%, 4.8%, 11.2%, 16.3%, and 19.7%, respectively (
Figure 5).
As the CO
2 ratio increased, the proportion of type X methanotrophs increased accordingly, suggesting that CO
2 was closely related to the proliferation of type X methanotrophs.
Table 2 shows the metabolite products (methanol, formaldehyde, formate) and coenzyme ratio (NADH/NAD
+) of methanotrophs at CH
4/CO
2 ratios of 0.5, 1.0, 1.5, 2.0, and 2.5. As shown in
Figure 5, a higher CO
2 ratio was associated with increased distribution of type X methanotrophs. In contrast, the metabolites and NADH/NAD
+ ratio of methanotrophs decreased as CO
2 ratio increased. Additionally, as CO
2 ratio increased, the proportion of type X methanotrophs increased, whereas the total number of methanotrophs decreased (the ratio of methanotrophs in the total bacterial community decreased to 48.3%, 46.3%, 41.2%, 36.5%, and 31.9% at CO
2/CH
4 ratios of 0.5, 1.0, 1.5, 2.0, and 2.5, respectively). Thus, as the total number of methanotrophs decreased, methanol production was impaired.
These findings suggest that gas composition ratio is an important factor affecting the efficiency of methanol production by methanotrophs. Moreover, it was thought reasonable to apply type X methanotrophs for the biological fixation and utilization of CO2 rather than for increasing the efficiency of methanol production. Type X methanotrophs also produce methanol by oxidizing methane via pMMO; however, the methane-oxidizing capacity of type X methanotrophs could not be compared to that of other methanotroph strains, as the methanotrophs were separately cultured. In subsequent experiments, we indirectly evaluated the CO2 utilization characteristics of type X methanotrophs for methanol production in the absence of methane.
2.3. Characteristics of CO2 Utilization by Methanotrophs through the Methane Oxidation Process
Residual concentrations of CH
4 and CO
2 in the flask headspace were measured to indirectly estimate the utilization rates of CH
4 and CO
2 with reaction time. The initial condition of culture medium and biogas was maintained, not changed every 24 h. The relative distribution of Type X methanotrophs strains was highest in the microbial sludge where cultured cells were centrifuged and dehydrated under the conditions of CH
4:CO
2 ratio 2.5, as shown in
Figure 5. The bacteria community was inoculated with 864 mg∙L
−1 based on the mixed liquid suspended solid (MLSS; Activated Bacteria Sludge). In the initial 0–24 hr stage, the CO
2 consumption rate was −1.32 mL CO
2·day
−1, which inversely increased the CO
2 content in the flask headspace. In the 72–96 h stage, as the CH
4 consumption rate was significantly lowered from the initial 5.30 to 0.14 mL CH
4·day
−1, the CO
2 consumption rate increased up to 2.37 mL CO
2·day
−1. This implies that CO
2 was consumed at the same reducing equivalent as NADH formed in the methane oxidation process by Type X after methane was depleted. In this study, although the changes in CH
4 and CO
2 utilization rates with reaction time was not monitored all time, the difference in the consumption rate of the two gases was evident in
Table 3. The initial increase in CO
2 in the head space may have been due to bacteria′s respiration, which indirectly showed that the consumption rate of each gas was different, depending on the elapsed reaction, when methane and CO
2 coexisted. The CO
2 in the flask headspace rather increases due to the respiration of bacteria. Despite the initial CO
2 content of 31.3%, the CO
2 content of the biogas decreased to 22.6% after 96 h. From this result, the negative CO
2 balance can be estimated by Type X methanotrophs.
Based on these findings, gas composition ratio according to reaction time was established to favor methane oxidation in the first half of the operation time, as well as to reduce and remove CO2 in the second half using reducing equivalents (NADH, PHB) formed through methane oxidation.
After completion of methane oxidation by methanotrophs, reducing equivalents accumulated in bacterial cells as a source of CO
2. Reducing equivalents were quantitatively confirmed from the amount of methanol produced. After methane oxidation reaction at a CH
4/CO
2/O
2 ratio of 1:0.5:1, bacterial cells were centrifuged, as described in
Section 2.1. Next, 835 mg∙L
−1 MLSS of dehydrated microbial sludge was added to the AMS medium. Subsequently, only CO
2 and O
2 (1:1), without CH
4, were supplied into the head space for assessment of methanol production (
Figure 6), which was then compared with that under control conditions using biogas. The NAD accumulated in bacterial cells existed in two forms: the oxidized and reduced forms [abbreviated as NAD
+ and NADH (H for hydrogen), respectively; NAD
+ + Formate⇔NADH.
In the control group cultured with biogas containing methane, methanol concentration increased to a maximum of 5.34 mM at 48 h and gradually decreased afterwards. However, under culture with CO2 only, methanol concentration in the culture medium increased to 0.12 mM within 24 h and then decreased rapidly. As methanol was formed at a low concentration, the reducing equivalents (NADH, PHB) in the bacterial cells were insufficient for type X methanotrophs to reduce CO2. To confirm whether CO2 reduction by type X methanotrophs can be accelerated with sufficient levels of reducing equivalents, 1 mM formate was added to form NADHs as electron donors from NAD+. At 24 h after the addition of formate, methanol concentration increased significantly to 1.32 mM. This suggested that certain methanotrophs, such as type X methanotrophs, in the bacterial community cultured with biogas might fix and use CO2 if reducing equivalents were continuously supplied. However, the addition of electron donors, such as formate, is not economically effective. Thus, electron donors must be continuously recycled and reused within the system. In other words, for the biological fixation of CO2 by type X methanotrophs, methane must be present until formate is formed by the MMOs of methanotrophs. The optimal reactor operation conditions that enable continuous supply of the reducing equivalents required for CO2 reduction may allow the biological fixation and effective use of CO2 by type X methanotrophs. The concentration of bacterial cells cultured in AMS medium was 720–860 mg∙L−1. Accordingly, we artificially increased the concentration of bacterial cells to elevate the amount of available reducing equivalents in the cells to evaluate their effects. The characteristics of methanol production from CO2 reduction were determined. CO2 and O2 ratio was set to 1:1 using reducing equivalents, such as NADH, PHB, and NH2OH, which were accumulated in the reactor and cells by bacteria clusters in bacteria community.
According to the gas composition ratio shown in
Figure 5, the bacterial cell sludge cultured under each condition with different relative ratios of type X methanotrophs was collected, and methanol production was measured with increasing bacterial cell concentration in the culture medium.
Figure 7 shows the effects of an artificial increase in cell concentration on methanol production. As the concentration and relative ratio of type X methanotrophs increased, methanol production tended to increase. The maximum methanol production of 0.41 mM was achieved at type X methanotroph ratio of 19% and cell concentration of 1025 mg∙L
−1. This was slightly higher than the methanol production of 0.38 mM achieved by Patel et al. using a single species of
Methylosinus sporium cultured with CO
2 only [
19]. Taken together, these finding suggest that increasing bacterial cell concentration when methane oxidation is completed may be an efficient method for the biological fixation and effective use of CO
2.
2.4. Interactions and Roles of Bacterial Strains in the Biological Fixation and Effective Use of CO2
NAD
+ is an oxidizing agent that is reduced by accepting electrons from other molecules. Reduced NAD
+ is converted to NADH and can be used as a reducing agent that can donate electrons. This electron transfer reaction is the main function of nicotinamide adenine dinucleotide (NAD). NADH reduces another molecule (CO
2 in our study) and is re-oxidized to NAD
+. Thus, the reaction is reversible. This suggests that coenzymes are not consumed, and that NAD
+ and NADH can continuously alternate between the two forms [
36]. In our previous study, methane is oxidized by MMO to produce methanol, and methanol oxidation continues to form NADH when formate is finally converted to CO
2 by the formate dehydrogenase (FDH) enzyme [
36]. At this time, in addition to NADH, which was formed via oxidation by methanotrophs as a reducing equivalent that can be recycled within the system itself, the type II methanotrophs
M. parvus and
M. rosea also express sMMO even under high levels of copper, thereby generating PHB (
Figure 4). These
Methylocytis species help provide the necessary reducing equivalents. The PHB synthesized by these
Methylocytis species accumulate in the cells, making them efficient species for the generation of reducing equivalents [
35]. Ammonia-oxidizing bacteria use ammonia as an energy source and partially oxidize methane through the action of ammonia monooxygenase [
1,
31]. Ammonia monooxygenase level in ammonia-oxidizing bacteria is lower than MMO level owing to the competition between methane and ammonia, however, ammonia-oxidizing bacteria can oxidize methane to produce methanol [
37]. Hydroxylamine (NH
2OH) produced during ammonia oxidation by ammonia-oxidizing bacteria can also act as an electron donor [
31,
38]. When ammonia is present in the culture, NH
2OH is spontaneously formed as an electron donor during oxidation of ammonia by ammonia-oxidizing bacteria. When such a system is used in water treatment, nitrate and methanol produced at this point are sent to the anoxic condition denitrification reactor in the biological nitrogen removal process, in which they help the internally generated methanol to be used as a carbon source by denitrifier bacteria, contributing to the role of the generated methanol in the denitrification of nitrate to N
2.
As described in
Section 2.1,
Methylophilus strains that used the methanol produced by methanotrophs as an energy source had a high abundance. These strains also contributed to supplying NADH as a reducing equivalent required for CO
2 reduction in the process of methanol oxidization into CO
2 by
Methylophilus strains, which consumed nitrogen in the reaction system and produced methanol.
In this study, we isolated each strain, but could not quantitatively measure the biological fixation and usage of CO2 for each strain. However, on the basis of the bacterial clusters and strains observed in this experiment, we determined the roles and mechanisms of each cluster involved in methanol formation via CO2 reduction, namely the methanotrophs, Methylophilus, and ammonia-oxidizing bacteria clusters.
In a bioreaction catalytic reactor process using methane as a substrate, methane and CO
2 are greenhouse gases that cannot be used in the reaction system owing to their low solubility and mass transfer coefficient. The processes must be designed to prevent leakage and maximize the consumption of methane and CO
2 within the bioreactor by only supplying the consumed amount of gas by using a pressure sensor within the closed pressure bioreactor tank. Additional studies are needed to design and operate a reactor that can circulate the produced methane and CO
2. Previous studies employing the internal [
39] and external [
40] loop airlift bioreactor methods for the circulation and use of methane in the manufacture of effective substances, such as methanol, by methanotrophs may provide an important basis for the development of optimal reactors.
3. Materials and Methods
3.1. Cultivation of Bacterial Consortium
Activated sludge collected from an aeration tank in a wastewater treatment plant was used as a microbial consortium culture. Wet activated sludge was filtered through a sieve (No. 50; 300 μm) to remove large-sized substances and was placed in several 500-mL flasks. Biogas was injected into 30% of the upper space filled with air, and the product was stored at 4 °C for use in subsequent experiments. As ammonium was the material of interest in this study, AMS medium containing a high concentration (10 mM) of ammonium chloride was prepared in the laboratory and used as a nitrogen source instead of NMS medium, which is commonly used for culturing methanotrophs [
41]. Copper (CuSO
4) concentration in the final medium was 20 μM for all medium formulations. The media were buffered to pH 6.8 through the addition of 1.5 mL phosphate buffer (26 g/L KH
2PO
4, 33 g/L Na
2HPO
4).
Next, 10 g of purified activated sludge sample stored at 4 °C was added to a 350 mL flask containing 200 mL of AMS medium solution. The flask was closed with a silicon stopper, and 20% (v/v) of the upper 150 mL space was filled with methane through a gas-tight syringe. The flask was subsequently sealed using a silicone stopper, covered with parafilm, and cultured on a rotary shaker (Lab Champion IS-971R; Champion Laboratories, Albion, IL, USA). After stirring for 24 h at a culture temperature of 25 °C and stirring speed of 250 rpm, the mixture was allowed to precipitate for 10 min. Next, 100 mL of the mixture was placed in a 350 mL flask containing fresh AMS, and the culture process was repeated six times. The cultured methane-oxidizing bacteria were separated via centrifugation (Centrifuge-416; Dongseo Science, Ltd., Dangjin, Korea) at 2700× g. The centrifuged pellet was freeze-dried at −55 °C in a freeze dryer (FDS-12003; OPERON, Seoul, Korea) for subsequent use. Cell concentration was determined by measuring the absorbance at a wavelength of 660 nm using a UV-visible spectrophotometer (UV-1600; Shimazu, Japan). The centrifuged pellet was freeze-dried at −55 °C (TFD5505; Ilshin lab., Korea). Different amounts of distilled water were added according to the concentration, and absorbance was measured to prepare a calibration curve.
3.2. Biogas Source
Biogas generated from an anaerobic digester in a city wastewater treatment plant (Ilsan, Goyang city, Korea) was collected in multiple 4.9 L high-pressure gas tanks (GlobalGastec, Ltd., Bucheon, Korea) and transferred to the laboratory for subsequent experiments. The composition ratio of biogas was CH4 (66.9%), CO2 (31.3%), N2 (1.2%), and O2 (0.2%). When it was difficult to control the content of each gas with only biogas and air during the production of mixed gas, CH4 and CO2 were supplied to the biogas from a high-pressure gas tank.
3.3. Biogas Analysis
Biogas was analyzed using a gas chromatographer as previously described [
24]. A gas chromatographer (HP 6890A; Agilent, Santa Clara, CA, USA) equipped with a packed column (30 m × 0.32 mm, GS-GASPRO; Agilent, Santa Clara, CA, USA) and a flame ionization detector was used. The inlet temperature was 100 °C; the oven temperature was 50 °C (3 min); the detector temperature was 200 °C; the carrier gas was He; and the flow rate was set to 1.2 mL/min, with a split ratio of 20:1.
3.4. Measurement of Metabolites and Coenzymes
Methane content in the serum bottle headspace was measured using a gas chromatography system (HP 6890A; Agilent) equipped with a packed column (30 m × 0.32 mm, GS-GASPRO) and a flame ionization detector. The inlet temperature, oven temperature, and detector temperature were 100 °C, 50 °C (3 min), and 200 °C, respectively. He was used as the carrier gas. The flow rate and split ratio were 1.2 mL/min and 20:1, respectively.
Methanol was analyzed using a gas chromatography system (Agilent HP 6890A, head space G1888) equipped with a packed column (30 m × 0.25 mm × 0.25 μm, INNOWAX; Agilent, Santa Clara, CA, USA) and a flame ionization detector. The oven temperature was increased from 50°C (5 min) to 250 °C (3 min), and the detector temperature was 250 °C. N2 was used as the carrier gas. The flow rate and split ratio were 1.2 mL/min and 10:1, respectively.
Formaldehyde was analyzed using high-performance liquid chromatography (HPLC) (Agilent 1100 series) equipped with a column (5 µm, 4.6 × 150 mm, Eclipse XDB-C18; Agilent, Santa Clara, CA, USA). The mobile phase consisted of A (D.W 100%) and B (ACN 100%). The gradient elution was performed at an A:B ratio of 50:50, flow rate of 1.5 mL/min, run time of 38 min, and UV detection of 360 nm.
Formate was analyzed using an HPLC system (Agilent 1100 series) equipped with a column (8 μm, 7.7 × 300 mm, Agilent Hi-Plex H). The injection volume was 20 µL. The mobile phase, flow rate, eluent, run time, and detection wavelength were 0.01 M 100% H2SO4, 0.6 mL/min, 0.01 M 100% H2SO4, 60 min, and 210 nm, respectively.
Residual NADH/ NAD+ in bacterial cells in the community were measured using the NAD+, NADH Colorimetric Assay Kit (Promega, Madison, WI, USA). The NADH-selective marker develops color upon reaction with NADH and can be detected at an optical density of 450 nm. To make an NADH standard solution (1 mM, 1 nmol/µL), 200 µL of 1X phosphate-buffered solution (PBS) was added into a vial containing the NADH standard (Component C). To prepare an NAD/NADH working solution, 8 mL of NADH Probe Buffer (Component B-II) was added to a bottle containing the NAD/NADH Recycling Enzyme Mix (Component A); next, to the bottle containing a mixture of Component A and B-II, 2 mL of NADH Probe (Component B-I) was added and mixed well.
3.5. Analysis of Microbial Community
DNA extraction, PCR amplifications, and pyrosequencing for microbial analysis were performed by ChunLab Inc. (
http://www.chunlab.com; Seoul, Korea). The 16S rRNA genes were amplified using barcoded universal primers for each sample. The conditions for each step of touch-down PCR are shown in
Table 4.
3.6. Microbiome and Genome Data Analysis
To compare operational taxonomic units between microbiome, genome, and microbial population samples analyzed by PCR and pyrosequencing, shared operational taxonomic units between the samples were identified through XOR analysis by ChunLab Inc. The composition and ratio of bacteria shared in the sample set were calculated.
4. Conclusions
This study propose the use of biogas generated from an urban wastewater treatment plant and a bacterial community comprising methanotrophs, Methylophilus, and ammonia-oxidizing bacteria, as an effective method of biological carbon fixation and nitrogen removal in wastewater treatment for CCUS. A significant level (11.2%) of type X methanotroph strains were identified only in methanotrophs cultured with CO2 and biogas. Following culture with CO2 only, higher cell concentration of bacterial community and greater relative ratio of type X methanotrophs were observed with increasing methanol production. At type X methanotroph ratio of 19% and cell concentration of 1025 mg∙L−1, the maximum methanol concentration of 0.41 mM was achieved. The most ideal reaction in bacteria community was determined in this study. Biological fixation of CO2 by type X methanotrophs was occurred using reducing equivalents, such as NADH, PHB, and NH2OH, which were accumulated in the reactor and cells by methanotrophs (Type I, Type II, and Type X), methylophilus and ammonia-oxidizing bacteria. Based on these findings, it would be possible to set a response system with conditions favorable for the oxidation of methane, methanol, and ammonia in the first half of the operation time and to reduce CO2 using reducing equivalents (NADH, PHB) for biological fixation in the second half of the operation time.