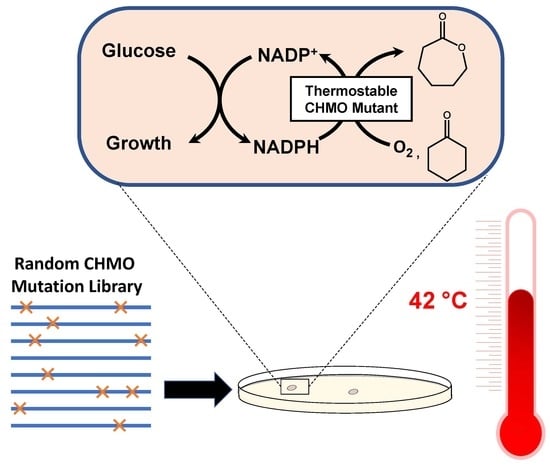
In Vivo, High-Throughput Selection of Thermostable Cyclohexanone Monooxygenase (CHMO)
Abstract
:1. Introduction
2. Results
2.1. Identification of CHMO Variants with Increased Thermostability
2.2. The Effects of Thermostability Enhancing Mutations on Protein Dynamics
2.3. Further Improving Thermostablity by Rational Design
2.4. Cyclohexanone to Caprolactone Conversion Using Thermostable CHMO
3. Discussions
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kim, T.H.; Kang, S.H.; Han, J.E.; Seo, E.J.; Jeon, E.Y.; Choi, G.E.; Park, J.B.; Oh, D.K. Multilayer Engineering of Enzyme Cascade Catalysis for One-Pot Preparation of Nylon Monomers from Renewable Fatty Acids. ACS Catal. 2020, 4871–4878. [Google Scholar] [CrossRef]
- Seo, J.H.; Kim, H.H.; Jeon, E.Y.; Song, Y.H.; Shin, C.S.; Park, J.B. Engineering of Baeyer-Villiger Monooxygenase-Based Escherichia Coli Biocatalyst for Large Scale Biotransformation of Ricinoleic Acid into (Z)-11-(Heptanoyloxy)Undec-9-Enoic Acid. Sci. Rep. 2016, 6, 28223. [Google Scholar] [CrossRef] [PubMed]
- Balke, K.; Bäumgen, M.; Bornscheuer, U.T. Controlling the Regioselectivity of Baeyer–Villiger Monooxygenases by Mutation of Active-Site Residues. ChemBioChem 2017, 18, 1627–1638. [Google Scholar] [CrossRef] [PubMed]
- Zambianchi, F.; Pasta, P.; Carrea, G.; Colonna, S.; Gaggero, N.; Woodley, J.M. Use of Isolated Cyclohexanone Monooxygenase from Recombinant Escherichia Coli as a Biocatalyst for Baeyer-Villiger and Sulfide Oxidations. Biotechnol. Bioeng. 2002, 78, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Walton, A.Z.; Stewart, J.D. Understanding and Improving NADPH-Dependent Reactions by Nongrowing Escherichia Coli Cells. Biotechnol. Prog. 2008, 20, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Romero, E.; Castellanos, J.R.G.; Mattevi, A.; Fraaije, M.W. Characterization and Crystal Structure of a Robust Cyclohexanone Monooxygenase. Angew. Chemie Int. Ed. 2016, 55, 15852–15855. [Google Scholar] [CrossRef]
- Rehdorf, J.; Kirschner, A.; Bornscheuer, U.T. Cloning, Expression and Characterization of a Baeyer-Villiger Monooxygenase from Pseudomonas Putida KT2440. Biotechnol. Lett. 2007, 29, 1393–1398. [Google Scholar] [CrossRef]
- Bisagni, S.; Abolhalaj, M.; de Brevern, A.G.; Rebehmed, J.; Hatti-Kaul, R.; Mamo, G. Enhancing the Activity of a Dietzia Sp. D5 Baeyer-Villiger Monooxygenase towards Cyclohexanone by Saturation Mutagenesis. ChemistrySelect 2017, 2, 7169–7177. [Google Scholar] [CrossRef]
- Schmidt, S.; Genz, M.; Balke, K.; Bornscheuer, U.T. The Effect of Disulfide Bond Introduction and Related Cys/Ser Mutations on the Stability of a Cyclohexanone Monooxygenase. J. Biotechnol. 2015, 214, 199–211. [Google Scholar] [CrossRef]
- van Beek, H.L.; Wijma, H.J.; Fromont, L.; Janssen, D.B.; Fraaije, M.W. Stabilization of Cyclohexanone Monooxygenase by a Computationally Designed Disulfide Bond Spanning Only One Residue. FEBS Open Bio 2014, 4, 168–174. [Google Scholar] [CrossRef] [Green Version]
- van Beek, H.L.; de Gonzalo, G.; Fraaije, M.W. Blending Baeyer-Villiger Monooxygenases: Using a Robust BVMO as a Scaffold for Creating Chimeric Enzymes with Novel Catalytic Properties. Chem. Commun. 2012, 48, 3288–3290. [Google Scholar] [CrossRef] [PubMed]
- Eijsink, V.G.H.; Bjørk, A.; Gåseidnes, S.; Sirevåg, R.; Synstad, B.; Van Den Burg, B.; Vriend, G. Rational Engineering of Enzyme Stability. J. Biotechnol. 2004, 113, 105–120. [Google Scholar] [CrossRef] [PubMed]
- Wijma, H.J.; Floor, R.J.; Janssen, D.B. Structure- and Sequence-Analysis Inspired Engineering of Proteins for Enhanced Thermostability. Curr. Opin. Struct. Biol. 2013, 23, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Soh, L.M.J.; Mak, W.S.; Lin, P.P.; Mi, L.; Chen, F.Y.H.; Damoiseaux, R.; Siegel, J.B.; Liao, J.C. Engineering a Thermostable Keto Acid Decarboxylase Using Directed Evolution and Computationally Directed Protein Design. ACS Synth. Biol. 2017, 6, 610–618. [Google Scholar] [CrossRef]
- Huang, R.; Chen, H.; Zhou, W.; Ma, C.; Zhang, Y.H.P. Engineering a Thermostable Highly Active Glucose 6-Phosphate Dehydrogenase and Its Application to Hydrogen Production in Vitro. Appl. Microbiol. Biotechnol. 2018, 102, 3203–3215. [Google Scholar] [CrossRef]
- Zhang, X.F.; Yang, G.Y.; Zhang, Y.; Xie, Y.; Withers, S.G.; Feng, Y. A General and Efficient Strategy for Generating the Stable Enzymes. Sci. Rep. 2016, 6, 33797. [Google Scholar] [CrossRef]
- Maxel, S.; Aspacio, D.; King, E.; Zhang, L.; Acosta, A.P.; Li, H. A Growth-Based, High-Throughput Selection Platform Enables Remodeling of 4-Hydroxybenzoate Hydroxylase Active Site. ACS Catal. 2020, 10, 6969–6974. [Google Scholar] [CrossRef]
- Machado, H.B.; Dekishima, Y.; Luo, H.; Lan, E.I.; Liao, J.C. A Selection Platform for Carbon Chain Elongation Using the CoA-Dependent Pathway to Produce Linear Higher Alcohols. Metab. Eng. 2012, 14, 504–511. [Google Scholar] [CrossRef]
- Liang, K.; Shen, C.R. Selection of an Endogenous 2,3-Butanediol Pathway in Escherichia Coli by Fermentative Redox Balance. Metab. Eng. 2017, 39, 181–191. [Google Scholar] [CrossRef]
- Zhang, L.; King, E.; Luo, R.; Li, H. Development of a High-Throughput, In Vivo Selection Platform for NADPH-Dependent Reactions Based on Redox Balance Principles. ACS Synth. Biol. 2018, 7, 1715–1721. [Google Scholar] [CrossRef]
- Calzadiaz-Ramirez, L.; Calvó-Tusell, C.; Stoffel, G.M.M.; Lindner, S.N.; Osuna, S.; Erb, T.J.; Garcia-Borràs, M.; Bar-Even, A.; Acevedo-Rocha, C.G. In Vivo Selection for Formate Dehydrogenases with High Efficiency and Specificity toward NADP +. ACS Catal. 2020, 7512–7525. [Google Scholar] [CrossRef] [PubMed]
- Kramer, L.; Le, X.; Rodriguez, M.; Wilson, M.A.; Guo, J.; Niu, W. Engineering Carboxylic Acid Reductase (CAR) through A Whole-Cell Growth-Coupled NADPH Recycling Strategy. ACS Synth. Biol. 2020, 9, 1632–1637. [Google Scholar] [CrossRef]
- Romero, P.A.; Arnold, F.H. Exploring Protein Fitness Landscapes by Directed Evolution. Nat. Rev. Mol. Cell Biol. 2009, 10, 866–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirza, I.A.; Yachnin, B.J.; Wang, S.; Grosse, S.; Imura, A.; Iwaki, H.; Hasegawa, Y. Crystal Structures of Cyclohexanone Monooxygenase Reveal Complex Domain Movements and a Sliding Cofactor. J. Am. Chem. Soc. 2009, 15, 8848–8854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Robinson, A.C.; Van Dam, M.E.; Martinez, P.; Economou, C.; Arnold, F.H. Enzyme Engineering for Nonaqueous Solvents. II. Additive Effects of Mutations on the Stability and Activity of Subtilisin E in Polar Organic Media. Biotechnol. Prog. 1991, 7, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Black, W.B.; Zhang, L.; Mak, W.S.; Maxel, S.; Cui, Y.; King, E.; Fong, B.; Sanchez Martinez, A.; Siegel, J.B.; Li, H. Engineering a Nicotinamide Mononucleotide Redox Cofactor System for Biocatalysis. Nat. Chem. Biol. 2020, 16, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Modarres, H.P.; Mofrad, M.R.; Sanati-Nezhad, A. Protein Thermostability Engineering. RSC Adv. 2016, 6, 115252–115270. [Google Scholar] [CrossRef]
- Borgo, B.; Havranek, J.J. Automated Selection of Stabilizing Mutations in Designed and Natural Proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 1494–1499. [Google Scholar] [CrossRef] [Green Version]
- Dill, K.A.; Ozkan, S.B.; Shell, M.S.; Weikl, T.R. The Protein Folding Problem. Annu. Rev. Biophys. 2008, 37, 289–316. [Google Scholar] [CrossRef]
- Yu, H.; Yan, Y.; Zhang, C.; Dalby, P.A. Two Strategies to Engineer Flexible Loops for Improved Enzyme Thermostability. Sci. Rep. 2017, 7, 41212. [Google Scholar] [CrossRef] [Green Version]
- Otten, R.; Liu, L.; Kenner, L.R.; Clarkson, M.W.; Mavor, D.; Tawfik, D.S.; Kern, D.; Fraser, J.S. Rescue of Conformational Dynamics in Enzyme Catalysis by Directed Evolution. Nat. Commun. 2018, 9, 1314. [Google Scholar] [CrossRef] [PubMed]
- Johansson, K.E.; Johansen, N.T.; Christensen, S.; Horowitz, S.; Bardwell JC, A.; Olsen, J.G.; Willemoës, M.; Lindorff-Larsen, K.; Ferkinghoff-Borg, J.; Hamelryck, T.; et al. Computational Redesign of Thioredoxin Is Hypersensitive toward Minor Conformational Changes in the Backbone Template. J. Mol. Biol. 2016, 428, 4361–4377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, C.H.; Chang PI, F.L.; Yeh, K.W.; Lin, W.C.; Chen, Y.M.; Lin, C.Y. Expression of a Gene Encoding a 16.9-KDa Heat-Shock Protein, Oshsp16.9, in Escherichia Coli Enhances Thermotolerance. Proc. Natl. Acad. Sci. USA 1997, 94, 10967–10972. [Google Scholar] [CrossRef] [Green Version]
- Rudolph, B.; Gebendorfer, K.M.; Buchner, J.; Winter, J. Evolution of Escherichia Coli for Growth at High Temperatures. J. Biol. Chem. 2010, 285, 19029–19034. [Google Scholar] [CrossRef] [Green Version]
- Lin, P.P.; Rabe, K.S.; Takasumi, J.L.; Kadisch, M.; Arnold, F.H.; Liao, J.C. Isobutanol Production at Elevated Temperatures in Thermophilic Geobacillus Thermoglucosidasius. Metab. Eng. 2014, 24, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datsenko, K.A.; Wanner, B.L. One-Step Inactivation of Chromosomal Genes in Escherichia Coli K-12 Using PCR Products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleishman, S.J.; Leaver-Fay, A.; Corn, J.E.; Strauch, E.M.; Khare, S.D.; Koga, N.; Ashworth, J.; Murphy, P.; Richter, F.; Lemmon, G.; et al. Rosettascripts: A Scripting Language Interface to the Rosetta Macromolecular Modeling Suite. PLoS ONE 2011, 6, e20161. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Dimaio, F.; Wang, R.Y.R.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-Resolution Comparative Modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef] [Green Version]
- Madden, T. Chapter 16: The BLAST Sequence Analysis Tool; National Center for Biotechnology Information: Bethesda, MD, USA, 2002.
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Yachnin, B.J.; Sprules, T.; McEvoy, M.B.; Lau PC, K.; Berghuis, A.M. The Substrate-Bound Crystal Structure of a Baeyer-Villiger Monooxygenase Exhibits a Criegee-like Conformation. J. Am. Chem. Soc. 2012, 134, 7788–7795. [Google Scholar] [CrossRef]
- Yachnin, B.J.; McEvoy, M.B.; Maccuish RJ, D.; Morley, K.L.; Lau PC, K.; Berghuis, A.M. Lactone-Bound Structures of Cyclohexanone Monooxygenase Provide Insight into the Stereochemistry of Catalysis. ACS Chem. Biol. 2014, 9, 2843–2851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber Biomolecular Simulation Programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [Green Version]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Antony, J.; Medvedev, D.M.; Stuchebrukhov, A.A. Theoretical Study of Electron Transfer between the Photolyase Catalytic Cofactor FADH- and DNA Thymine Dimer. J. Am. Chem. Soc. 2000, 122, 1057–1065. [Google Scholar] [CrossRef]
- Walker, R.C.; De Souza, M.M.; Mercer, I.P.; Gould, I.R.; Klug, D.R. Large and Fast Relaxations inside a Protein: Calculation and Measurement of Reorganization Energies in Alcohol Dehydrogenase. J. Phys. Chem. B 2002, 106, 11658–11665. [Google Scholar] [CrossRef] [Green Version]
- Anandakrishnan, R.; Aguilar, B.; Onufriev, A.V. H++ 3.0: Automating pK Prediction and the Preparation of Biomolecular Structures for Atomistic Molecular Modeling and Simulations. Nucleic Acids Res. 2012, 40, 537–541. [Google Scholar] [CrossRef] [Green Version]
- Ringnér, M. What Is Principal Component Analysis? Nat. Biotechnol. 2008, 26, 303–304. [Google Scholar] [CrossRef]
- Bahmani, B.; Moseley, B.; Vattani, A.; Kumar, R.; Vassilvitskii, S. Scalable κ-Means++. Proc. VLDB Endow. 2012, 5, 622–633. [Google Scholar] [CrossRef]
- Alford, R.F.; Leaver-Fay, A.; Jeliazkov, J.R.; O’Meara, M.J.; DiMaio, F.P.; Park, H.; Shapovalov, M.V.; Renfrew, P.D.; Mulligan, V.K.; Kappel, K.; et al. The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design. J. Chem. Theory Comput. 2017, 13, 3031–3048. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Vanderplas, J.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar] [CrossRef]
- Van Der Walt, S.; Colbert, S.C.; Varoquaux, G. The NumPy Array: A Structure for Efficient Numerical Computation. Comput. Sci. Eng. 2011, 13, 22–30. [Google Scholar] [CrossRef] [Green Version]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- McGibbon, R.T.; Beauchamp, K.A.; Harrigan, M.P.; Klein, C.; Swails, J.M.; Hernández, C.X.; Schwantes, C.R.; Wang, L.P.; Lane, T.J.; Pande, V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015, 109, 1528–1532. [Google Scholar] [CrossRef] [Green Version]
- The PyMOL Molecular Graphics System, Version 1.8; Schrödinger, LLC: New York, NY, USA, 2015.





| NADPH | Cyclohexanone | |||||
|---|---|---|---|---|---|---|
| Enzymes | KM (µM) | kcat (s−1) | kcat/KM (µM−1s−1) | KM (µM) | kcat (s−1) | kcat/KM (µM−1s−1) |
| Wild type | 36.6 ± 8.36 | 13.5 ± 0.55 | 0.37 | 4.09 ± 1.81 | 12.9 ± 0.86 | 3.15 |
| GV | 43.3 ± 20.9 | 9.67 ± 1.23 | 0.22 | 5.64 ± 0.48 | 11.6 ± 1.55 | 2.05 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maxel, S.; Zhang, L.; King, E.; Acosta, A.P.; Luo, R.; Li, H. In Vivo, High-Throughput Selection of Thermostable Cyclohexanone Monooxygenase (CHMO). Catalysts 2020, 10, 935. https://doi.org/10.3390/catal10080935
Maxel S, Zhang L, King E, Acosta AP, Luo R, Li H. In Vivo, High-Throughput Selection of Thermostable Cyclohexanone Monooxygenase (CHMO). Catalysts. 2020; 10(8):935. https://doi.org/10.3390/catal10080935
Chicago/Turabian StyleMaxel, Sarah, Linyue Zhang, Edward King, Ana Paula Acosta, Ray Luo, and Han Li. 2020. "In Vivo, High-Throughput Selection of Thermostable Cyclohexanone Monooxygenase (CHMO)" Catalysts 10, no. 8: 935. https://doi.org/10.3390/catal10080935