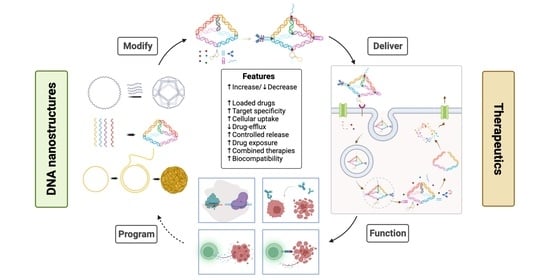
Therapeutic Applications of Programmable DNA Nanostructures
Abstract
:1. Introduction
2. Fate of DNA Nanostructures in Living Cells
2.1. Targeted Drug Delivery
2.2. Cellular Uptake of DNA Nanostructures
2.3. Stability of DNA Nanostructures in Physiological Conditions
3. Molecular Payloads
3.1. Doxorubicin
| Structure | Payload | Modification | Results | Ref. |
|---|---|---|---|---|
| TDN | DOX | Folate receptor | 6 h incubation induced apoptosis of HT 29 colon cancer cells. | [59] |
| TDN | HApt | Enhanced stability and prolonged circulation of HApt, induced apoptosis and arrested cell growth. | [66] | |
| TDN | DOX | Affibody | Bind ~ 53 molecules of DOX with greater selectivity and inhibition of breast cancer cells. | [67] |
| TDN | DOX | Folate receptor | A synergic anti-cancer biological effect with chemotherapy. | [59] |
| TDN | 5-FU | AS1411 aptamer | Better targeting ability to kill breast cancer. | [68] |
| TDN | DOX | AS1411 + MUC1 aptamer | Lower cytotoxicity to MUC1-negative cells, equal lethality to sensitive cells, and more effective to DOX resistant cells. | [69] |
| TDN | TMZ | AS1411 + GS24 | Attenuate drug resistance in temozolomide (TMZ)-resistant cells with specific binding to transferrin receptor. | [70] |
| TDN | Ir | AS1411 + MUC1 aptamer | Inhibits the growth and invasion of glioma cells. | [71] |
| TDN | ASOs | Nuclear localization peptide | Antisense strands released inhibit cell proliferation at a low concentration without transfection reagent with efficient knockdown of the c-raf gene. | [72] |
| TDN | DOX | Efficient delivery of DOX into drug-resistant breast cancer cells. | [64] | |
| TDN | DOX | KLA peptide | 3KLA-modified TDNs designed for mitochondrial targeting exhibited the most efficient DOX accumulation in mitochondria of 4T1 cells leading to an effective release of cytochrome c, and the upregulated expression levels of caspase-9, caspase-3, p21, and p53. | [73] |
| NF | DOX | Circumvent drug-resistant cells with less side effects to non-target cells. | [63] | |
| NF | DOX | Sgc8 | Preparation of multifunctional DNA Nanoflowers that are resistant to nuclease and can integrate with different functional moieties. | [74] |
| DNA triangle | DOX | Exhibited remarkable anti-tumor efficacy without systemic toxicity in mice with orthotopic breast tumors. | [51] | |
| DNA triangle | BMEPC | Cellular-level dual-functional imaging and photodynamic therapy that generates free radicals and subsequent apoptosis. | [75] | |
| DNA triangle and tube | DOX | Increased cellular internalization of DOX with enhanced cell-killing activity to drug-resistant adenocarcinoma cells. | [55] | |
| DNA tube with conformational change to DNA sheet | Thrombin | AS1411 aptamer | Nucleolin-targeting aptamer serves both as a targeting domain and as a molecular trigger for the mechanical opening of DNA nanorobot delivering thrombin, specifically tumor-associated blood vessels, and inducing intravascular thrombosis resulting in tumor necrosis and inhibition of tumor growth. | [76] |
| DNA icosahedron | DOX | MUC1 aptamer | DOX@Apt-DNA-icosa shows efficient and specific internalization for killing epithelial cancer cells. | [77] |
| DNA dendrimer | EPI | AS1411+ MUC1 aptamer | Apts-Dendrimer-Epi complex released Epi in a pH-sensitive manner (more release at pH 5.5), prohibiting tumor growth in vitro and in vivo. | [78] |
| DNA nanorod | Daunorub-icin | Circumvent efflux pump-mediated drug resistance in leukemia cells at clinically relevant drug concentrations. | [65] | |
| DNA nanocircuit | Chlorin e6 | Aptamer | Aptamer-based DNA nanocircuit selectively recognizes target cancer cells, activates photosensitizers, and amplifies the photodynamic therapeutic effect. | [79] |
| DNA nanotrain | DOX | AS1411, Sgc8 | Locomotives guiding nanotrains with boxcars carrying high payload allowing intracellular signaling. | [80] |
| DNA nanocentipede | DOX | Zy1 | Effective binding affinity and selectivity with enhanced cellular cytotoxicity to the target cell but not to negative control cells. | [61] |
| X-Y-Shaped DNA | DOX | Sgc8 | Specific cytotoxic effect against leukemia cells with the incorporation of therapeutic antisense oligonucleotides inhibiting efflux pump of drug circumventing drug resistance. | [81] |
| Biotinylated octahedral DNA nanocages | DOX | Folic acid | DOX-loaded Bio-Fol-DNA nanocages delivered DOX selectively to the folate receptor-enriched Hela cells. | [82] |
| A 3D tubular DNA origami with six helical bundles | DOX | DUPA (a small molecule ligand) | Ligand conjugate DONs delivered DOX with high affinity and selectivity into the prostate-specific membrane antigen (PSMA)+ cancer cell line LNCaP. DOX-DUPA-DONs showed lower toxicity against PC-3 cells (PSMA-) in comparison to free DOX. | [83] |
| tFNA (Tetral framework nucleic acid) | Maytansin-e (DM1) | HApt-aptamer | HApt-tFNA@DM1 (HApDC) could target HER2 protein and delivered chemotherapeutic agents into HER2-positive breast tumor. HApDCs exerted enhanced anti-tumor efficiency in comparison with free drug and synthetic liposome-derived vesicles without side effects. | [84] |
| All-sealed divalent aptamer Tetrahedral DNA framework (asdTDF) | Therapeutic protein | Aptamer | The ligase-assisted seal of the nicks resulted in the enhanced TDF stability against nuclease digestion protecting the therapeutic protein from degradation. Endogenous gluathione can trigger the release of therapeutic protein leading to the apoptosis of the specific cancer cells. | [85] |
| Tetrahedral DNA | Photother-anostic molecule (IR780) | The in vitro and ex vivo photothermal and photodynamic efficiencies of IR780 in the tumor site was high in IR780@Td with enhanced tumor imaging and anti-tumor effects than IR780 alone. | [86] | |
| A triplex-DNA nanoswitch | Drug combo (Antisense DNA, Cisplatin, DOX | Aptamer | The effects of gene silencing and significant inhibition of tumor growth was shown with tumor-bearing mouse models upon intravenous administration of smart pH responsive nanoswitch that can be used for combinatorial cancer therapy. | [87] |
3.2. Therapeutic Nucleic Acid Delivery
3.3. Delivery of Gene Editing Tools
4. DNA Nanorobots That Deliver Molecular Payloads
5. Tetrahedral Framework Nucleic Acids as Therapeutic Agents
6. DNA Nanostructures Interacting with the Cell Membrane
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ghosh, S.; Banerjee, M. A Smart Viral Vector for Targeted Delivery of Hydrophobic Drugs. Sci. Rep. 2021, 11, 7030. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, Y.; Wei, D.; Singh, P.; Yu, Y.; Lee, T.; Zhang, L.; Mandl, H.K.; Piotrowski-Daspit, A.S.; Chen, X.; et al. Nanoparticle-Mediated Convection-Enhanced Delivery of a DNA Intercalator to Gliomas Circumvents Temozolomide Resistance. Nat. Biomed. Eng. 2021, 5, 1048–1058. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.K.; Hafez, I.M.; Baoukina, S.; Belliveau, N.M.; Zhigaltsev, I.V.; Afshinmanesh, E.; Tieleman, D.P.; Hansen, C.L.; Hope, M.J.; Cullis, P.R. Lipid Nanoparticles Containing SiRNA Synthesized by Microfluidic Mixing Exhibit an Electron-Dense Nanostructured Core. J. Phys. Chem. C 2012, 116, 18440–18450. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Qin, J.; Jiang, Y.; Duncan, R.G.; Brigham, B.; Fishman, S.; Nair, J.K.; Akinc, A.; Barros, S.A.; Kasperkovitz, P.V. Shielding of Lipid Nanoparticles for SiRNA Delivery: Impact on Physicochemical Properties, Cytokine Induction, and Efficacy. Mol. Ther. Nucleic Acids 2014, 3, e210. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Marioli, M.; Zhang, K. Analytical Characterization of Liposomes and Other Lipid Nanoparticles for Drug Delivery. J. Pharm. Biomed. Anal. 2021, 192, 113642. [Google Scholar] [CrossRef] [PubMed]
- Graczyk, A.; Pawlowska, R.; Jedrzejczyk, D.; Chworos, A. Gold Nanoparticles in Conjunction with Nucleic Acids as a Modern Molecular System for Cellular Delivery. Molecules 2020, 25, 204. [Google Scholar] [CrossRef] [Green Version]
- Roma-Rodrigues, C.; Pereira, F.; Alves de Matos, A.P.; Fernandes, M.; Baptista, P.V.; Fernandes, A.R. Smuggling Gold Nanoparticles across Cell Types—A New Role for Exosomes in Gene Silencing. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1389–1398. [Google Scholar] [CrossRef]
- Huang, X.; El-Sayed, M.A. Gold Nanoparticles: Optical Properties and Implementations in Cancer Diagnosis and Photothermal Therapy. J. Adv. Res. 2010, 1, 13–28. [Google Scholar] [CrossRef] [Green Version]
- Seeman, N.C. Nucleic Acid Junctions and Lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Winfree, E.; Liu, F.; Wenzler, L.A.; Seeman, N.C. Design and Self-Assembly of Two-Dimensional DNA Crystals. Nature 1998, 394, 539–544. [Google Scholar] [CrossRef]
- Rothemund, P.W.K. Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy-Nissenbaum, E.; Radovic-Moreno, A.F.; Wang, A.Z.; Langer, R.; Farokhzad, O.C. Nanotechnology and Aptamers: Applications in Drug Delivery. Trends Biotechnol. 2008, 26, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Godonoga, M.; Lin, T.-Y.; Oshima, A.; Sumitomo, K.; Tang, M.S.L.; Cheung, Y.-W.; Kinghorn, A.B.; Dirkzwager, R.M.; Zhou, C.; Kuzuya, A.; et al. A DNA Aptamer Recognising a Malaria Protein Biomarker Can Function as Part of a DNA Origami Assembly. Sci. Rep. 2016, 6, 21266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arap, W.; Pasqualini, R.; Ruoslahti, E. Cancer Treatment by Targeted Drug Delivery to Tumor Vasculature in a Mouse Model. Science 1998, 279, 377–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, M.; Wang, Y.; Zhao, W.; Qi, R.; Han, Y.; Wu, R.; Wang, Y.; Xu, H. Peptide-Induced DNA Condensation into Virus-Mimicking Nanostructures. ACS Appl. Mater. Interfaces 2018, 10, 24349–24360. [Google Scholar] [CrossRef]
- Schrama, D.; Reisfeld, R.A.; Becker, J.C. Antibody Targeted Drugs as Cancer Therapeutics. Nat. Rev. Drug. Discov. 2006, 5, 147–159. [Google Scholar] [CrossRef]
- Ranallo, S.; Prévost-Tremblay, C.; Idili, A.; Vallée-Bélisle, A.; Ricci, F. Antibody-Powered Nucleic Acid Release Using a DNA-Based Nanomachine. Nat. Commun. 2017, 8, 15150. [Google Scholar] [CrossRef] [Green Version]
- Sudimack, J.; Lee, R.J. Targeted Drug Delivery via the Folate Receptor. Adv. Drug Deliv. Rev. 2000, 41, 147–162. [Google Scholar] [CrossRef]
- Ko, S.; Liu, H.; Chen, Y.; Mao, C. DNA Nanotubes as Combinatorial Vehicles for Cellular Delivery. Biomacromolecules 2008, 9, 3039–3043. [Google Scholar] [CrossRef] [Green Version]
- Walsh, A.S.; Yin, H.; Erben, C.M.; Wood, M.J.A.; Turberfield, A.J. DNA Cage Delivery to Mammalian Cells. ACS Nano 2011, 5, 5427–5432. [Google Scholar] [CrossRef]
- Hamblin, G.D.; Carneiro, K.M.M.; Fakhoury, J.F.; Bujold, K.E.; Sleiman, H.F. Rolling Circle Amplification-Templated DNA Nanotubes Show Increased Stability and Cell Penetration Ability. J. Am. Chem. Soc. 2012, 134, 2888–2891. [Google Scholar] [CrossRef] [PubMed]
- Giljohann, D.A.; Seferos, D.S.; Patel, P.C.; Millstone, J.E.; Rosi, N.L.; Mirkin, C.A. Oligonucleotide Loading Determines Cellular Uptake of DNA-Modified Gold Nanoparticles. Nano Lett. 2007, 7, 3818–3821. [Google Scholar] [CrossRef] [PubMed]
- Dix, J.A.; Verkman, A.S. Crowding Effects on Diffusion in Solutions and Cells. Annu. Rev. Biophys. 2008, 37, 247–263. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Li, J.; Li, Q.; Huang, Q.; Shi, J.; Yan, H.; Fan, C. Single-Particle Tracking and Modulation of Cell Entry Pathways of a Tetrahedral DNA Nanostructure in Live Cells. Angew. Chem. Int. Ed. 2014, 53, 7745–7750. [Google Scholar] [CrossRef]
- Vale, R.D.; Milligan, R.A. The Way Things Move: Looking Under the Hood of Molecular Motor Proteins. Science 2000, 288, 88–95. [Google Scholar] [CrossRef] [Green Version]
- Soldati, T.; Schliwa, M. Powering Membrane Traffic in Endocytosis and Recycling. Nat. Rev. Mol. Cell. Biol. 2006, 7, 897–908. [Google Scholar] [CrossRef]
- Nichols, B.J.; Lippincott-Schwartz, J. Endocytosis without Clathrin Coats. Trends Cell Biol. 2001, 11, 406–412. [Google Scholar] [CrossRef]
- Mikkilä, J.; Eskelinen, A.-P.; Niemelä, E.H.; Linko, V.; Frilander, M.J.; Törmä, P.; Kostiainen, M.A. Virus-Encapsulated DNA Origami Nanostructures for Cellular Delivery. Nano Lett. 2014, 14, 2196–2200. [Google Scholar] [CrossRef] [Green Version]
- Perrault, S.D.; Shih, W.M. Virus-Inspired Membrane Encapsulation of DNA Nanostructures To Achieve In Vivo Stability. ACS Nano 2014, 8, 5132–5140. [Google Scholar] [CrossRef]
- Sanchez-Rueda, E.G.; Rodriguez-Cristobal, E.; González, C.L.M.; Hernandez-Garcia, A. Protein-Coated DsDNA Nanostars with High Structural Rigidity and High Enzymatic and Thermal Stability. Nanoscale 2019, 11, 18604–18611. [Google Scholar] [CrossRef]
- Auvinen, H.; Zhang, H.; Nonappa; Kopilow, A.; Niemelä, E.H.; Nummelin, S.; Correia, A.; Santos, H.A.; Linko, V.; Kostiainen, M.A. Protein Coating of DNA Nanostructures for Enhanced Stability and Immunocompatibility. Adv. Healthc. Mater. 2017, 6, 1700692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez-Garcia, A. Strategies to Build Hybrid Protein-DNA Nanostructures. Nanomaterials 2021, 11, 1332. [Google Scholar] [CrossRef] [PubMed]
- Hahn, J.; Wickham, S.F.J.; Shih, W.M.; Perrault, S.D. Addressing the Instability of DNA Nanostructures in Tissue Culture. ACS Nano 2014, 8, 8765–8775. [Google Scholar] [CrossRef]
- Keum, J.-W.; Bermudez, H. Enhanced Resistance of DNA Nanostructures to Enzymatic Digestion. Chem. Commun. 2009, 45, 7036–7038. [Google Scholar] [CrossRef]
- Mei, Q.; Wei, X.; Su, F.; Liu, Y.; Youngbull, C.; Johnson, R.; Lindsay, S.; Yan, H.; Meldrum, D. Stability of DNA Origami Nanoarrays in Cell Lysate. Nano Lett. 2011, 11, 1477–1482. [Google Scholar] [CrossRef] [Green Version]
- Castro, C.E.; Kilchherr, F.; Kim, D.-N.; Shiao, E.L.; Wauer, T.; Wortmann, P.; Bathe, M.; Dietz, H. A Primer to Scaffolded DNA Origami. Nat. Methods 2011, 8, 221–229. [Google Scholar] [CrossRef]
- Goltry, S.; Hallstrom, N.; Clark, T.; Kuang, W.; Lee, J.; Jorcyk, C.; Knowlton, W.B.; Yurke, B.; Hughes, W.L.; Graugnard, E. DNA Topology Influences Molecular Machine Lifetime in Human Serum. Nanoscale 2015, 7, 10382–10390. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, A.R.; Vilcapoma, J.; Dey, P.; Wong-Deyrup, S.W.; Dey, B.K.; Halvorsen, K. Exceptional Nuclease Resistance of Paranemic Crossover (PX) DNA and Crossover-Dependent Biostability of DNA Motifs. J. Am. Chem. Soc. 2020, 142, 6814–6821. [Google Scholar] [CrossRef]
- Li, Y.; Song, L.; Wang, B.; He, J.; Li, Y.; Deng, Z.; Mao, C. Universal PH-Responsive and Metal-Ion-Free Self-Assembly of DNA Nanostructures. Angew. Chem. Int. Ed. 2018, 57, 6892–6895. [Google Scholar] [CrossRef]
- Cassinelli, V.; Oberleitner, B.; Sobotta, J.; Nickels, P.; Grossi, G.; Kempter, S.; Frischmuth, T.; Liedl, T.; Manetto, A. One-Step Formation of “Chain-Armor”-Stabilized DNA Nanostructures. Angew. Chem. Int. Ed. 2015, 54, 7795–7798. [Google Scholar] [CrossRef]
- Gerling, T.; Kube, M.; Kick, B.; Dietz, H. Sequence-Programmable Covalent Bonding of Designed DNA Assemblies. Sci. Adv. 2018, 4, eaau1157. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Ke, Y.; Li, Z.; Wang, J.H.; Liu, Y.; Yan, H. Mirror Image DNA Nanostructures for Chiral Supramolecular Assemblies. Nano Lett. 2009, 9, 433–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacroix, A.; Vengut-Climent, E.; de Rochambeau, D.; Sleiman, H.F. Uptake and Fate of Fluorescently Labeled DNA Nanostructures in Cellular Environments: A Cautionary Tale. ACS Cent. Sci. 2019, 5, 882–891. [Google Scholar] [CrossRef] [Green Version]
- Conway, J.W.; McLaughlin, C.K.; Castor, K.J.; Sleiman, H. DNA Nanostructure Serum Stability: Greater than the Sum of Its Parts. Chem. Commun. 2013, 49, 1172–1174. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Yin, P. Enhancing Biocompatible Stability of DNA Nanostructures Using Dendritic Oligonucleotides and Brick Motifs. Angew. Chem. Int. Ed. 2020, 59, 700–703. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, Y.; Llano, E.D.; Barišić, I. (Poly)Cation-Induced Protection of Conventional and Wireframe DNA Origami Nanostructures. Nanoscale 2018, 10, 7494–7504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponnuswamy, N.; Bastings, M.M.C.; Nathwani, B.; Ryu, J.H.; Chou, L.Y.T.; Vinther, M.; Li, W.A.; Anastassacos, F.M.; Mooney, D.J.; Shih, W.M. Oligolysine-Based Coating Protects DNA Nanostructures from Low-Salt Denaturation and Nuclease Degradation. Nat. Commun. 2017, 8, 15654. [Google Scholar] [CrossRef]
- Chandrasekaran, A.R.; Halvorsen, K. Nuclease Degradation Analysis of DNA Nanostructures Using Gel Electrophoresis. Curr. Protoc. Nucleic Acid Chem. 2020, 82, e115. [Google Scholar] [CrossRef]
- Liu, X.; Xu, Y.; Yu, T.; Clifford, C.; Liu, Y.; Yan, H.; Chang, Y. A DNA Nanostructure Platform for Directed Assembly of Synthetic Vaccines. Nano Lett. 2012, 12, 4254–4259. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Song, L.; Liu, S.; Jiang, Q.; Liu, Q.; Li, N.; Wang, Z.-G.; Ding, B. A DNA-Based Nanocarrier for Efficient Gene Delivery and Combined Cancer Therapy. Nano Lett. 2018, 18, 3328–3334. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, Q.; Li, N.; Dai, L.; Liu, Q.; Song, L.; Wang, J.; Li, Y.; Tian, J.; Ding, B.; et al. DNA Origami as an In Vivo Drug Delivery Vehicle for Cancer Therapy. ACS Nano 2014, 8, 6633–6643. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Ji, W.; Hall, J.M.; Hu, Q.; Wang, C.; Beisel, C.L.; Gu, Z. Self-Assembled DNA Nanoclews for the Efficient Delivery of CRISPR–Cas9 for Genome Editing. Angew. Chem. Int. Ed. 2015, 54, 12029–12033. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Hu, R.; Zhao, Z.; Chen, Z.; Zhang, X.; Tan, W. Noncanonical Self-Assembly of Multifunctional DNA Nanoflowers for Biomedical Applications. J. Am. Chem. Soc. 2013, 135, 16438–16445. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, N.; Wu, J.; Xu, X.; Kamaly, N.; Farokhzad, O.C. Cancer Nanotechnology: The Impact of Passive and Active Targeting in the Era of Modern Cancer Biology. Adv. Drug Deliv. Rev. 2014, 66, 2–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Q.; Song, C.; Nangreave, J.; Liu, X.; Lin, L.; Qiu, D.; Wang, Z.-G.; Zou, G.; Liang, X.; Yan, H.; et al. DNA Origami as a Carrier for Circumvention of Drug Resistance. J. Am. Chem. Soc. 2012, 134, 13396–13403. [Google Scholar] [CrossRef]
- Zhao, Y.-X.; Shaw, A.; Zeng, X.; Benson, E.; Nyström, A.M.; Högberg, B. DNA Origami Delivery System for Cancer Therapy with Tunable Release Properties. ACS Nano 2012, 6, 8684–8691. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.H.; Kim, K.-R.; Lee, H.; Ahn, D.-R.; Ko, Y.T. In Vitro and in Vivo Behavior of DNA Tetrahedrons as Tumor-Targeting Nanocarriers for Doxorubicin Delivery. Colloids Surf. B Biointerfaces 2017, 157, 424–431. [Google Scholar] [CrossRef]
- Liu, M.; Ma, W.; Li, Q.; Zhao, D.; Shao, X.; Huang, Q.; Hao, L.; Lin, Y. Aptamer-Targeted DNA Nanostructures with Doxorubicin to Treat Protein Tyrosine Kinase 7-Positive Tumours. Cell Prolif. 2019, 52, e12511. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Zhang, Z.; Yang, J. DNA Tetrahedron Delivery Enhances Doxorubicin-Induced Apoptosis of HT-29 Colon Cancer Cells. Nanoscale Res. Lett. 2017, 12, 495. [Google Scholar] [CrossRef] [Green Version]
- Sun, P.; Zhang, N.; Tang, Y.; Yang, Y.; Chu, X.; Zhao, Y. SL2B Aptamer and Folic Acid Dual-Targeting DNA Nanostructures for Synergic Biological Effect with Chemotherapy to Combat Colorectal Cancer. Int. J. Nanomed. 2017, 12, 2657–2672. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Yang, X.; He, L.; Wang, K.; Wang, Q.; Huang, J.; Liu, J.; Wu, B.; Xu, C. Self-Assembled DNA Nanocentipede as Multivalent Drug Carrier for Targeted Delivery. ACS Appl. Mater. Interfaces 2016, 8, 25733–25740. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.M.; Schindler, M. Cell Biological Mechanisms of Multidrug Resistance in Tumors. Proc. Natl. Acad. Sci. USA 1994, 91, 3497–3504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, L.; Zhu, G.; Qiu, L.; Wu, C.; Chen, H.; Liang, H.; Cansiz, S.; Lv, Y.; Zhang, X.; Tan, W. Self-Assembled Multifunctional DNA Nanoflowers for the Circumvention of Multidrug Resistance in Targeted Anticancer Drug Delivery. Nano Res. 2015, 8, 3447–3460. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-R.; Kim, D.-R.; Lee, T.; Yhee, J.Y.; Kim, B.-S.; Kwon, I.C.; Ahn, D.-R. Drug Delivery by a Self-Assembled DNA Tetrahedron for Overcoming Drug Resistance in Breast Cancer Cells. Chem. Commun. 2013, 49, 2010–2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halley, P.D.; Lucas, C.R.; McWilliams, E.M.; Webber, M.J.; Patton, R.A.; Kural, C.; Lucas, D.M.; Byrd, J.C.; Castro, C.E. Daunorubicin-Loaded DNA Origami Nanostructures Circumvent Drug-Resistance Mechanisms in a Leukemia Model. Small 2016, 12, 308–320. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Zhan, Y.; Zhang, Y.; Shao, X.; Xie, X.; Mao, C.; Cui, W.; Li, Q.; Shi, J.; Li, J.; et al. An Intelligent DNA Nanorobot with in Vitro Enhanced Protein Lysosomal Degradation of HER. Nano Lett. 2019, 19, 4505–4517. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Jiang, S.; Zhang, D.; Bai, X.M.; Hecht, S.; Chen, S. DNA–Affibody Nanoparticles for Inhibiting Breast Cancer Cells Overexpressing HER. Chem. Commun. 2017, 53, 573–576. [Google Scholar] [CrossRef]
- Zhan, Y.; Ma, W.; Zhang, Y.; Mao, C.; Shao, X.; Xie, X.; Wang, F.; Liu, X.; Li, Q.; Lin, Y. DNA-Based Nanomedicine with Targeting and Enhancement of Therapeutic Efficacy of Breast Cancer Cells. ACS Appl. Mater. Interfaces 2019, 11, 15354–15365. [Google Scholar] [CrossRef]
- Liu, X.; Wu, L.; Wang, L.; Jiang, W. A Dual-Targeting DNA Tetrahedron Nanocarrier for Breast Cancer Cell Imaging and Drug Delivery. Talanta 2018, 179, 356–363. [Google Scholar] [CrossRef]
- Fu, W.; You, C.; Ma, L.; Li, H.; Ju, Y.; Guo, X.; Shi, S.; Zhang, T.; Zhou, R.; Lin, Y. Enhanced Efficacy of Temozolomide Loaded by a Tetrahedral Framework DNA Nanoparticle in the Therapy for Glioblastoma. ACS Appl. Mater. Interfaces 2019, 11, 39525–39533. [Google Scholar] [CrossRef]
- Tian, Y.; Huang, Y.; Gao, P.; Chen, T. Nucleus-Targeted DNA Tetrahedron as a Nanocarrier of Metal Complexes for Enhanced Glioma Therapy. Chem. Commun. 2018, 54, 9394–9397. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Jiang, Q.; He, L.; Zhan, P.; Liu, Q.; Liu, S.; Fu, M.; Liu, J.; Li, C.; Ding, B. Self-Assembled Double-Bundle DNA Tetrahedron for Efficient Antisense Delivery. ACS Appl. Mater. Interfaces 2018, 10, 23693–23699. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Chen, J.; Zhang, N.; Yang, Y.; Zhu, W.; Li, L.; He, B. Mitochondria-Targeted Tetrahedral DNA Nanostructures for Doxorubicin Delivery and Enhancement of Apoptosis. J. Mater. Chem. B 2020, 8, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Hu, R.; Zhu, G.; Zhang, X.; Mei, L.; Liu, Q.; Qiu, L.; Wu, C.; Tan, W. Preparation and Biomedical Applications of Programmable and Multifunctional DNA Nanoflowers. Nat. Protoc. 2015, 10, 1508–1524. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, X.; Ma, X.; Xue, X.; Jiang, Q.; Song, L.; Dai, L.; Zhang, C.; Jin, S.; Yang, K.; Ding, B.; et al. A Photosensitizer-Loaded DNA Origami Nanosystem for Photodynamic Therapy. ACS Nano 2016, 10, 3486–3495. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Jiang, Q.; Liu, S.; Zhang, Y.; Tian, Y.; Song, C.; Wang, J.; Zou, Y.; Anderson, G.J.; Han, J.-Y.; et al. A DNA Nanorobot Functions as a Cancer Therapeutic in Response to a Molecular Trigger in Vivo. Nat. Biotechnol. 2018, 36, 258–264. [Google Scholar] [CrossRef]
- Chang, M.; Yang, C.-S.; Huang, D.-M. Aptamer-Conjugated DNA Icosahedral Nanoparticles As a Carrier of Doxorubicin for Cancer Therapy. ACS Nano 2011, 5, 6156–6163. [Google Scholar] [CrossRef]
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Lavaee, P.; Jalalian, S.H.; Robati, R.Y.; Abnous, K. Double Targeting and Aptamer-Assisted Controlled Release Delivery of Epirubicin to Cancer Cells by Aptamers-Based Dendrimer in Vitro and in Vivo. Eur. J. Pharm. Biopharm. 2016, 102, 152–158. [Google Scholar] [CrossRef]
- Han, D.; Zhu, G.; Wu, C.; Zhu, Z.; Chen, T.; Zhang, X.; Tan, W. Engineering a Cell-Surface Aptamer Circuit for Targeted and Amplified Photodynamic Cancer Therapy. ACS Nano 2013, 7, 2312–2319. [Google Scholar] [CrossRef]
- Zhu, G.; Zheng, J.; Song, E.; Donovan, M.; Zhang, K.; Liu, C.; Tan, W. Self-Assembled, Aptamer-Tethered DNA Nanotrains for Targeted Transport of Molecular Drugs in Cancer Theranostics. Proc. Natl. Acad. Sci. USA 2013, 110, 7998–8003. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Han, D.; Chen, T.; Peng, L.; Zhu, G.; You, M.; Qiu, L.; Sefah, K.; Zhang, X.; Tan, W. Building a Multifunctional Aptamer-Based DNA Nanoassembly for Targeted Cancer Therapy. J. Am. Chem. Soc. 2013, 135, 18644–18650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raniolo, S.; Vindigni, G.; Ottaviani, A.; Unida, V.; Iacovelli, F.; Manetto, A.; Figini, M.; Stella, L.; Desideri, A.; Biocca, S. Selective Targeting and Degradation of Doxorubicin-Loaded Folate-Functionalized DNA Nanocages. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1181–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, Z.; Guo, L.; Wu, G.; Li, J.; Sun, Y.; Hou, Y.; Shi, J.; Song, S.; Wang, L.; Fan, C.; et al. DNA Origami-Enabled Engineering of Ligand–Drug Conjugates for Targeted Drug Delivery. Small 2020, 16, 1904857. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Yang, Y.; Zhu, J.; Jia, W.; Zhang, T.; Liu, Z.; Chen, X.; Lin, Y. Biomimetic Nanoerythrosome-Coated Aptamer-DNA Tetrahedron/Maytansine Conjugates: PH-Responsive and Targeted Cytotoxicity for HER2-Positive Breast Cancer. Adv. Mater. 2022. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, X.; Yang, F.; Yuan, R.; Xiang, Y. Targeted Delivery of DNA Framework-Encapsulated Native Therapeutic Protein into Cancer Cells. ACS Appl. Mater. Interfaces 2020, 12, 54489–54496. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, Z.; Tong, Y.; Zhai, Y.; Zhao, X.; Yue, X.; Qiao, Y.; Liu, Y.; Yin, Y.; Xi, R.; et al. Improved Cancer Phototheranostic Efficacy of Hydrophobic IR780 via Parenteral Route by Association with Tetrahedral Nanostructured DNA. J. Control. Release 2021, 330, 483–492. [Google Scholar] [CrossRef]
- Chen, X.; Chen, T.; Ren, L.; Chen, G.; Gao, X.; Li, G.; Zhu, X. Triplex DNA Nanoswitch for PH-Sensitive Release of Multiple Cancer Drugs. ACS Nano 2019, 13, 7333–7344. [Google Scholar] [CrossRef]
- Krieg, A.M.; Yi, A.-K.; Matson, S.; Waldschmidt, T.J.; Bishop, G.A.; Teasdale, R.; Koretzky, G.A.; Klinman, D.M. CpG Motifs in Bacterial DNA Trigger Direct B-Cell Activation. Nature 1995, 374, 546–549. [Google Scholar] [CrossRef]
- Hemmi, H.; Takeuchi, O.; Kawai, T.; Kaisho, T.; Sato, S.; Sanjo, H.; Matsumoto, M.; Hoshino, K.; Wagner, H.; Takeda, K.; et al. A Toll-like Receptor Recognizes Bacterial DNA. Nature 2000, 408, 740–745. [Google Scholar] [CrossRef]
- Schmidt, M.; Anton, K.; Nordhaus, C.; Junghans, C.; Wittig, B.; Worm, M. Cytokine and Ig-Production by CG-Containing Sequences with Phosphorodiester Backbone and Dumbbell-Shape. Allergy 2006, 61, 56–63. [Google Scholar] [CrossRef]
- Nishikawa, M.; Matono, M.; Rattanakiat, S.; Matsuoka, N.; Takakura, Y. Enhanced Immunostimulatory Activity of Oligodeoxynucleotides by Y-Shape Formation. Immunology 2008, 124, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Rattanakiat, S.; Nishikawa, M.; Funabashi, H.; Luo, D.; Takakura, Y. The Assembly of a Short Linear Natural Cytosine-Phosphate-Guanine DNA into Dendritic Structures and Its Effect on Immunostimulatory Activity. Biomaterials 2009, 30, 5701–5706. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tseng, Y.D.; Kwon, S.Y.; d’Espaux, L.; Bunch, J.S.; McEuen, P.L.; Luo, D. Controlled Assembly of Dendrimer-like DNA. Nat. Mater. 2004, 3, 38–42. [Google Scholar] [CrossRef]
- Ouyang, X.; Li, J.; Liu, H.; Zhao, B.; Yan, J.; Ma, Y.; Xiao, S.; Song, S.; Huang, Q.; Chao, J.; et al. Rolling Circle Amplification-Based DNA Origami Nanostructrures for Intracellular Delivery of Immunostimulatory Drugs. Small 2013, 9, 3082–3087. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, G.; Mei, L.; Wu, C.; Qiu, L.; Cui, C.; Liu, Y.; Teng, I.-T.; Tan, W. Self-Assembled DNA Immunonanoflowers as Multivalent CpG Nanoagents. ACS Appl. Mater. Interfaces 2015, 7, 24069–24074. [Google Scholar] [CrossRef]
- Mohri, K.; Nishikawa, M.; Takahashi, N.; Shiomi, T.; Matsuoka, N.; Ogawa, K.; Endo, M.; Hidaka, K.; Sugiyama, H.; Takahashi, Y.; et al. Design and Development of Nanosized DNA Assemblies in Polypod-like Structures as Efficient Vehicles for Immunostimulatory CpG Motifs to Immune Cells. ACS Nano 2012, 6, 5931–5940. [Google Scholar] [CrossRef]
- Li, J.; Pei, H.; Zhu, B.; Liang, L.; Wei, M.; He, Y.; Chen, N.; Li, D.; Huang, Q.; Fan, C. Self-Assembled Multivalent DNA Nanostructures for Noninvasive Intracellular Delivery of Immunostimulatory CpG Oligonucleotides. ACS Nano 2011, 5, 8783–8789. [Google Scholar] [CrossRef]
- Schüller, V.J.; Heidegger, S.; Sandholzer, N.; Nickels, P.C.; Suhartha, N.A.; Endres, S.; Bourquin, C.; Liedl, T. Cellular Immunostimulation by CpG-Sequence-Coated DNA Origami Structures. ACS Nano 2011, 5, 9696–9702. [Google Scholar] [CrossRef] [Green Version]
- Charoenphol, P.; Bermudez, H. Aptamer-Targeted DNA Nanostructures for Therapeutic Delivery. Mol. Pharm. 2014, 11, 1721–1725. [Google Scholar] [CrossRef]
- Lee, H.; Lytton-Jean, A.K.R.; Chen, Y.; Love, K.T.; Park, A.I.; Karagiannis, E.D.; Sehgal, A.; Querbes, W.; Zurenko, C.S.; Jayaraman, M.; et al. Molecularly Self-Assembled Nucleic Acid Nanoparticles for Targeted in Vivo SiRNA Delivery. Nat. Nanotechnol. 2012, 7, 389–393. [Google Scholar] [CrossRef]
- Kim, K.-R.; Jegal, H.; Kim, J.; Ahn, D.-R. A Self-Assembled DNA Tetrahedron as a Carrier for in Vivo Liver-Specific Delivery of SiRNA. Biomater. Sci. 2020, 8, 586–590. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Ding, F.; Zhang, J.; Guo, Y.; Gao, X.; Feng, J.; Zhu, X.; Zhang, C. DNA Tetrahedron-Based Nanogels for SiRNA Delivery and Gene Silencing. Chem. Commun. 2019, 55, 4222–4225. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Chen, T.; Song, Y.; Feng, C.; Chen, H.; Zhang, Q.; Chen, G.; Zhu, X. MRNA Delivery by a PH-Responsive DNA Nano-Hydrogel. Small 2021, 17, e2101224. [Google Scholar] [CrossRef] [PubMed]
- Fakhoury, J.J.; McLaughlin, C.K.; Edwardson, T.W.; Conway, J.W.; Sleiman, H.F. Development and Characterization of Gene Silencing DNA Cages. Biomacromolecules 2014, 15, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Bujold, K.E.; Hsu, J.C.C.; Sleiman, H.F. Optimized DNA “Nanosuitcases” for Encapsulation and Conditional Release of SiRNA. J. Am. Chem. Soc. 2016, 138, 14030–14038. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Seelig, G. Dynamic DNA Nanotechnology Using Strand-Displacement Reactions. Nat. Chem. 2011, 3, 103–113. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Wu, T.; Lu, X.; Wu, X.; Liu, S.; Zhao, S.; Xu, X.; Ding, B. A Self-Assembled Platform Based on Branched DNA for SgRNA/Cas9/Antisense Delivery. J. Am. Chem. Soc. 2019, 141, 19032–19037. [Google Scholar] [CrossRef]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-Assembly of DNA into Nanoscale Three-Dimensional Shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef]
- Hamaguchi, N.; Ellington, A.; Stanton, M. Aptamer Beacons for the Direct Detection of Proteins. Anal. Biochem. 2001, 294, 126–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boniface, J.J.; Rabinowitz, J.D.; Wülfing, C.; Hampl, J.; Reich, Z.; Altman, J.D.; Kantor, R.M.; Beeson, C.; McConnell, H.M.; Davis, M.M. Initiation of Signal Transduction through the T Cell Receptor Requires the Multivalent Engagement of Peptide/MHC Ligands. Immunity 1998, 9, 459–466. [Google Scholar] [CrossRef] [Green Version]
- Amir, Y.; Ben-Ishay, E.; Levner, D.; Ittah, S.; Abu-Horowitz, A.; Bachelet, I. Universal Computing by DNA Origami Robots in a Living Animal. Nat. Nanotechnol. 2014, 9, 353–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Jiang, S.; Liu, X.; Pan, L.; Zhang, C. Aptamer-Binding Directed DNA Origami Pattern for Logic Gates. ACS Appl. Mater. Interfaces 2016, 8, 34054–34060. [Google Scholar] [CrossRef]
- Liu, F.; Liu, X.; Shi, Q.; Maffeo, C.; Kojima, M.; Dong, L.; Aksimentiev, A.; Huang, Q.; Fukuda, T.; Arai, T. A Tetrahedral DNA Nanorobot with Conformational Change in Response to Molecular Trigger. Nanoscale 2021, 13, 15552–15559. [Google Scholar] [CrossRef]
- Li, S.; Jiang, D.; Rosenkrans, Z.T.; Barnhart, T.E.; Ehlerding, E.B.; Ni, D.; Engle, J.W.; Cai, W. Aptamer-Conjugated Framework Nucleic Acids for the Repair of Cerebral Ischemia-Reperfusion Injury. Nano Lett. 2019, 19, 7334–7341. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, Q.; Wang, F.; Zhou, Z.; Xu, J.; Cheng, S.; Cheng, Y. Self-Assembled DNA Nanostructure as a Carrier for Targeted SiRNA Delivery in Glioma Cells. Int. J. Nanomed. 2021, 16, 1805–1817. [Google Scholar] [CrossRef]
- Li, F.; Lu, J.; Liu, J.; Liang, C.; Wang, M.; Wang, L.; Li, D.; Yao, H.; Zhang, Q.; Wen, J.; et al. A Water-Soluble Nucleolin Aptamer-Paclitaxel Conjugate for Tumor-Specific Targeting in Ovarian Cancer. Nat. Commun. 2017, 8, 1390. [Google Scholar] [CrossRef]
- Bates, P.J.; Reyes-Reyes, E.M.; Malik, M.T.; Murphy, E.M.; O’Toole, M.G.; Trent, J.O. G-Quadruplex Oligonucleotide AS1411 as a Cancer-Targeting Agent: Uses and Mechanisms. Biochim. Biophys. Acta-Gen. Subj. 2017, 1861, 1414–1428. [Google Scholar] [CrossRef]
- Shi, S.; Fu, W.; Lin, S.; Tian, T.; Li, S.; Shao, X.; Zhang, Y.; Zhang, T.; Tang, Z.; Zhou, Y.; et al. Targeted and Effective Glioblastoma Therapy via Aptamer-Modified Tetrahedral Framework Nucleic Acid-Paclitaxel Nanoconjugates That Can Pass the Blood Brain Barrier. Nanomed. Nanotechnol. Biol. Med. 2019, 21, 102061. [Google Scholar] [CrossRef]
- Xie, X.; Shao, X.; Ma, W.; Zhao, D.; Shi, S.; Li, Q.; Lin, Y. Overcoming Drug-Resistant Lung Cancer by Paclitaxel Loaded Tetrahedral DNA Nanostructures. Nanoscale 2018, 10, 5457–5465. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Zhan, Y.; Shao, X.; Fu, W.; Xiao, D.; Zhu, J.; Qin, X.; Zhang, T.; Zhang, M.; Zhou, Y.; et al. Neuroprotective and Neurotherapeutic Effects of Tetrahedral Framework Nucleic Acids on Parkinson’s Disease in Vitro. ACS Appl. Mater. Interfaces 2019, 11, 32787–32797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lin, S.; Wang, L.; Peng, S.; Tian, T.; Li, S.; Xiao, J.; Lin, Y. Tetrahedral Framework Nucleic Acids Act as Antioxidants in Acute Kidney Injury Treatment. Chem. Eng. J. 2021, 413, 127426. [Google Scholar] [CrossRef]
- Yao, Y.; Wen, Y.; Li, Y.; Zhu, J.; Tian, T.; Zhang, Q.; Xiao, D.; Gao, Y.; Lin, Y.; Wei, W.; et al. Tetrahedral Framework Nucleic Acids Facilitate Neurorestoration of Facial Nerves by Activating the NGF/PI3K/AKT Pathway. Nanoscale 2021, 13, 15598–15610. [Google Scholar] [CrossRef]
- Chen, X.; Xie, Y.; Liu, Z.; Lin, Y. Application of Programmable Tetrahedral Framework Nucleic Acid-Based Nanomaterials in Neurological Disorders: Progress and Prospects. Front. Bioeng. Biotechnol. 2021, 9, 782237. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, J.; Ma, W.; Zhang, W.; Xie, Y.; Chen, X.; Zhu, J.; Liu, Y.; Qin, X.; Lin, Y. The Remyelination Effect of DNA Framework Nucleic Acids on Demyelinating Diseases. Appl. Mater. Today 2021, 24, 101098. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, X.; Li, N.; Zhou, M.; Zhang, T.; Li, S.; Cai, X.; Ji, P.; Lin, Y. Tetrahedral Framework Nucleic Acids Promote Corneal Epithelial Wound Healing in Vitro and in Vivo. Small 2019, 15, 1901907. [Google Scholar] [CrossRef]
- Jiang, D.; Ge, Z.; Im, H.-J.; England, C.G.; Ni, D.; Hou, J.; Zhang, L.; Kutyreff, C.J.; Yan, Y.; Liu, Y.; et al. DNA Origami Nanostructures Can Exhibit Preferential Renal Uptake and Alleviate Acute Kidney Injury. Nat. Biomed. Eng. 2018, 2, 865–877. [Google Scholar] [CrossRef]
- Chen, Q.; Ding, F.; Zhang, S.; Li, Q.; Liu, X.; Song, H.; Zuo, X.; Fan, C.; Mou, S.; Ge, Z. Sequential Therapy of Acute Kidney Injury with a DNA Nanodevice. Nano Lett. 2021, 21, 4394–4402. [Google Scholar] [CrossRef]
- Li, S.; Liu, Y.; Tian, T.; Zhang, T.; Lin, S.; Zhou, M.; Zhang, X.; Lin, Y.; Cai, X. Bioswitchable Delivery of MicroRNA by Framework Nucleic Acids: Application to Bone Regeneration. Small 2021, 17, e2104359. [Google Scholar] [CrossRef]
- Qin, X.; Xiao, L.; Li, N.; Hou, C.; Li, W.; Li, J.; Yan, N.; Lin, Y. Tetrahedral Framework Nucleic Acids-Based Delivery of MicroRNA-155 Inhibits Choroidal Neovascularization by Regulating the Polarization of Macrophages. Bioact. Mater. 2021; in press. [Google Scholar] [CrossRef]
- Zhang, Y.; Xie, X.; Ma, W.; Zhan, Y.; Mao, C.; Shao, X.; Lin, Y. Multi-Targeted Antisense Oligonucleotide Delivery by a Framework Nucleic Acid for Inhibiting Biofilm Formation and Virulence. Nano-Micro Lett. 2020, 12, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Sun, Y.; Li, S.; Liu, M.; Qin, X.; Chen, X.; Lin, Y. Tetrahedral Framework Nucleic Acids Deliver Antimicrobial Peptides with Improved Effects and Less Susceptibility to Bacterial Degradation. Nano Lett. 2020, 20, 3602–3610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Tian, T.; Zhou, R.; Li, S.; Ma, W.; Zhang, Y.; Liu, N.; Shi, S.; Li, Q.; Xie, X.; et al. Design, Fabrication and Applications of Tetrahedral DNA Nanostructure-Based Multifunctional Complexes in Drug Delivery and Biomedical Treatment. Nat. Protoc. 2020, 15, 2728–2757. [Google Scholar] [CrossRef]
- Shao, X.; Ma, W.; Xie, X.; Li, Q.; Lin, S.; Zhang, T.; Lin, Y. Neuroprotective Effect of Tetrahedral DNA Nanostructures in a Cell Model of Alzheimer’s Disease. ACS Appl. Mater. Interfaces 2018, 10, 23682–23692. [Google Scholar] [CrossRef]
- Shao, X.; Cui, W.; Xie, X.; Ma, W.; Zhan, Y.; Lin, Y. Treatment of Alzheimer’s Disease with Framework Nucleic Acids. Cell Prolif. 2020, 53, e12787. [Google Scholar] [CrossRef]
- Qin, X.; Li, N.; Zhang, M.; Lin, S.; Zhu, J.; Xiao, D.; Cui, W.; Zhang, T.; Lin, Y.; Cai, X. Tetrahedral Framework Nucleic Acids Prevent Retina Ischemia-Reperfusion Injury from Oxidative Stress via Activating the Akt/Nrf2 Pathway. Nanoscale 2019, 11, 20667–20675. [Google Scholar] [CrossRef]
- Ma, W.; Zhan, Y.; Zhang, Y.; Xie, X.; Mao, C.; Lin, Y. Enhanced Neural Regeneration with a Concomitant Treatment of Framework Nucleic Acid and Stem Cells in Spinal Cord Injury. ACS Appl. Mater. Interfaces 2020, 12, 2095–2106. [Google Scholar] [CrossRef]
- Fu, W.; Ma, L.; Ju, Y.; Xu, J.; Li, H.; Shi, S.; Zhang, T.; Zhou, R.; Zhu, J.; Xu, R.; et al. Therapeutic SiCCR2 Loaded by Tetrahedral Framework DNA Nanorobotics in Therapy for Intracranial Hemorrhage. Adv. Funct. Mater. 2021, 31, 2101435. [Google Scholar] [CrossRef]
- Li, J.; Xiao, L.; Yan, N.; Li, Y.; Wang, Y.; Qin, X.; Zhao, D.; Liu, M.; Li, N.; Lin, Y. The Neuroprotective Effect of MicroRNA-22-3p Modified Tetrahedral Framework Nucleic Acids on Damaged Retinal Neurons Via TrkB/BDNF Signaling Pathway. Adv. Funct. Mater. 2021, 31, 2104141. [Google Scholar] [CrossRef]
- Cui, W.; Yang, X.; Chen, X.; Xiao, D.; Zhu, J.; Zhang, M.; Qin, X.; Ma, X.; Lin, Y. Treating LRRK2-Related Parkinson’s Disease by Inhibiting the MTOR Signaling Pathway to Restore Autophagy. Adv. Funct. Mater. 2021, 31, 2105152. [Google Scholar] [CrossRef]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. DNA Transfection Mediated by Cationic Lipid Reagents. Cold Spring Harb. Protoc. 2019, 2019, pdb.prot095414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spagnou, S.; Miller, A.D.; Keller, M. Lipidic Carriers of SiRNA: Differences in the Formulation, Cellular Uptake, and Delivery with Plasmid DNA. Biochemistry 2004, 43, 13348–13356. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Ren, G.; Simonsen, J.B.; Ryan, R.O. Cationic Lipid Nanodisks as an SiRNA Delivery Vehicle. Biochem. Cell Biol. 2014, 92, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Ewert, K.; Slack, N.L.; Ahmad, A.; Evans, H.M.; Lin, A.J.; Samuel, C.E.; Safinya, C.R. Cationic Lipid-DNA Complexes for Gene Therapy: Understanding the Relationship between Complex Structure and Gene Delivery Pathways at the Molecular Level. Curr. Med. Chem. 2004, 11, 133–149. [Google Scholar] [CrossRef] [Green Version]
- Ewert, K.; Ahmad, A.; Evans, H.M.; Safinya, C.R. Cationic Lipid–DNA Complexes for Non-Viral Gene Therapy: Relating Supramolecular Structures to Cellular Pathways. Expert Opin. Biol. Ther. 2005, 5, 33–53. [Google Scholar] [CrossRef]
- Sato, Y.; Endo, M.; Morita, M.; Takinoue, M.; Sugiyama, H.; Murata, S.; Nomura, S.M.; Suzuki, Y. Environment-Dependent Self-Assembly of DNA Origami Lattices on Phase-Separated Lipid Membranes. Adv. Mater. Interfaces 2018, 5, 1800437. [Google Scholar] [CrossRef]
- Burns, J.R.; Göpfrich, K.; Wood, J.W.; Thacker, V.V.; Stulz, E.; Keyser, U.F.; Howorka, S. Lipid-Bilayer-Spanning DNA Nanopores with a Bifunctional Porphyrin Anchor. Angew. Chem. Int. Ed. 2013, 52, 12069–12072. [Google Scholar] [CrossRef] [Green Version]
- Göpfrich, K.; Zettl, T.; Meijering, A.E.C.; Hernández-Ainsa, S.; Kocabey, S.; Liedl, T.; Keyser, U.F. DNA-Tile Structures Induce Ionic Currents through Lipid Membranes. Nano Lett. 2015, 15, 3134–3138. [Google Scholar] [CrossRef] [Green Version]
- Chidchob, P.; Offenbartl-Stiegert, D.; McCarthy, D.; Luo, X.; Li, J.; Howorka, S.; Sleiman, H.F. Spatial Presentation of Cholesterol Units on a DNA Cube as a Determinant of Membrane Protein-Mimicking Functions. J. Am. Chem. Soc. 2019, 141, 1100–1108. [Google Scholar] [CrossRef]
- Wang, M.; Zuris, J.A.; Meng, F.; Rees, H.; Sun, S.; Deng, P.; Han, Y.; Gao, X.; Pouli, D.; Wu, Q.; et al. Efficient Delivery of Genome-Editing Proteins Using Bioreducible Lipid Nanoparticles. Proc. Natl. Acad. Sci. USA 2016, 113, 2868–2873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Gao, Y.; Wang, Y.; Wei, Q.; Shi, J.; Chen, N.; Li, D.; Fan, C. Guiding Protein Delivery into Live Cells Using DNA-Programmed Membrane Fusion. Chem. Sci. 2018, 9, 5967–5975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, J.R.; Stulz, E.; Howorka, S. Self-Assembled DNA Nanopores That Span Lipid Bilayers. Nano Lett. 2013, 13, 2351–2356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, J.R.; Seifert, A.; Fertig, N.; Howorka, S. A Biomimetic DNA-Based Channel for the Ligand-Controlled Transport of Charged Molecular Cargo across a Biological Membrane. Nat. Nanotechnol. 2016, 11, 152–156. [Google Scholar] [CrossRef]
- Langecker, M.; Arnaut, V.; Martin, T.G.; List, J.; Renner, S.; Mayer, M.; Dietz, H.; Simmel, F.C. Synthetic Lipid Membrane Channels Formed by Designed DNA Nanostructures. Science 2012, 338, 6109. [Google Scholar] [CrossRef] [Green Version]
- Göpfrich, K.; Li, C.-Y.; Ricci, M.; Bhamidimarri, S.P.; Yoo, J.; Gyenes, B.; Ohmann, A.; Winterhalter, M.; Aksimentiev, A.; Keyser, U.F. Large-Conductance Transmembrane Porin Made from DNA Origami. ACS Nano 2016, 10, 8207–8214. [Google Scholar] [CrossRef]
- Göpfrich, K.; Li, C.-Y.; Mames, I.; Bhamidimarri, S.P.; Ricci, M.; Yoo, J.; Mames, A.; Ohmann, A.; Winterhalter, M.; Stulz, E.; et al. Ion Channels Made from a Single Membrane-Spanning DNA Duplex. Nano Lett. 2016, 16, 4665–4669. [Google Scholar] [CrossRef]
- Seifert, A.; Göpfrich, K.; Burns, J.R.; Fertig, N.; Keyser, U.F.; Howorka, S. Bilayer-Spanning DNA Nanopores with Voltage-Switching between Open and Closed State. ACS Nano 2015, 9, 1117–1126. [Google Scholar] [CrossRef] [Green Version]
- Maingi, V.; Burns, J.R.; Uusitalo, J.J.; Howorka, S.; Marrink, S.J.; Sansom, M.S.P. Stability and Dynamics of Membrane-Spanning DNA Nanopores. Nat. Commun. 2017, 8, 14784. [Google Scholar] [CrossRef] [Green Version]
- Fragasso, A.; De Franceschi, N.; Stömmer, P.; van der Sluis, E.O.; Dietz, H.; Dekker, C. Reconstitution of Ultrawide DNA Origami Pores in Liposomes for Transmembrane Transport of Macromolecules. ACS Nano 2021, 15, 12768–12779. [Google Scholar] [CrossRef]
- Kishi, K.; Yasuda, T.; Ikehara, Y.; Sawazaki, K.; Sato, W.; Iida, R. Human Serum Deoxyribonuclease I (DNase I) Polymorphism: Pattern Similarities among Isozymes from Serum, Urine, Kidney, Liver, and Pancreas. Am. J. Hum. Genet. 1990, 47, 121–126. [Google Scholar] [PubMed]
- Haran, G.; Cohen, R.; Bar, L.K.; Barenholz, Y. Transmembrane Ammonium Sulfate Gradients in Liposomes Produce Efficient and Stable Entrapment of Amphipathic Weak Bases. Biochim. Biophys. Acta Biomembr. 1993, 1151, 201–215. [Google Scholar] [CrossRef]
- Pound, E.; Ashton, J.R.; Becerril, H.A.; Woolley, A.T. Polymerase Chain Reaction Based Scaffold Preparation for the Production of Thin, Branched DNA Origami Nanostructures of Arbitrary Sizes. Nano Lett. 2009, 9, 4302–4305. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhang, C.; Zhang, C.; Zhao, Y.; Qi, Z.; Fan, C.; Chao, J.; Wei, B. DNA Origami Nanostructures with Scaffolds Obtained from Rolling Circle Amplification. ACS Mater. Lett. 2020, 2, 1322–1327. [Google Scholar] [CrossRef]
- Praetorius, F.; Kick, B.; Behler, K.L.; Honemann, M.N.; Weuster-Botz, D.; Dietz, H. Biotechnological Mass Production of DNA Origami. Nature 2017, 552, 84–87. [Google Scholar] [CrossRef]









| Structure | Size (nm) | Strategy | Test | Results | Ref. | |
|---|---|---|---|---|---|---|
| Before Modification | After Modification | |||||
| 24-HB | 100 | Close-packed helices | DNase I | Duplex plasmid DNA degraded in 5 min | Close-pack helices degraded in 1 h | [36] |
| Tweezers | 14 | Topology | 70% human serum | Open state in 20 h | Closed state in 37 h | [37] |
| Paranemic crossover (PX), Double crossover (DX), Duplex DNA | 13 | Increased crossovers | 10% FBS, human serum and urine, DNase I, Exonuclease V, T5 and T 7 | PX–not degraded DX–not degraded Duplex–degraded | [38] | |
| Octahedron | 50 | Heating FBS | Media + 10% FBS | 0% intact without heating | 100% intact with heating | [33] |
| Octahedron | 50 | Nuclease inhibitors | Media + 10% FBS | 0% intact without acting | 100% intact with actin | [33] |
| Nanotube | 400 | |||||
| Nanorod | 89 | |||||
| Tetrahedron | 14 | Ethylenediamine buffer | DNase I | 0% intact in TAE with Mg2+ buffer | 100% intact in ethylenediamine buffer | [39] |
| Nanotube | 30 | Crosslinking (Click chemistry) | Exonuclease | Fully degraded | Partially degraded for crosslinked | [40] |
| Brick-like DNA origami | 70 | Crosslinking (UV-induced T-T dimers) | DNase I | 10 min | 1 h | [41,42] |
| Triangular prism, tetrahedron | 7 | Hexanediol and hexamethylene glycol | Media + 10% FBS | 18 h lifetime | 55 h lifetime | [43,44] |
| DNA brick | 50 | Dendritic oligonucleotides | DNase I (100 U/mL) | Fully degraded with 5 U/mL | Coated— 50% degraded with 100 U/mL | [45] |
| Origami rod | 350 | Cationic polysaccharides | DNase I | Stable for 1 h | Stable for 24 h | [46] |
| Origami barrel | 60 | Oligolysine-PEG copolymer | Media + 10% FBS | 5 min half-life | 50 min half-life | [47] |
| Octahedron | 76 | PEGylated lipid bilayer | DNase I | 30% intact | 85% intact | [29] |
| 60 HB | 20× | BSA-dendron conjugates | Media + 10% FBS | 20% intact | 100% intact | [31] |
| 20× | ||||||
| 33 | ||||||
| 24 HB | 100 | Silica coating | DNase I | Completely degraded | Almost fully intact | [48] |
| Octahedron | 29 | Peptides | DNase I | Completely degraded | Almost fully intact | [48] |
| 4-Arm junction Nanotube | 5 30–70 | L-DNA (mirror form of D-DNA) | Exonuclease I Exonuclease III | Completely degraded | Almost fully intact | [42] |
| tFNA Design | Targeted Disease | Results | Ref. |
|---|---|---|---|
| tFNA with aptamer conjugation | Cerebral ischemia-reperfusion | Alleviate oxidative stress | [116] |
| tFNA-aptamer to deliver siRNA | Glioma cells | Apoptosis | [117] |
| tFNA | Alzheimer’s disease | Apoptosis | [135] |
| tFNA with aptamer and paclitaxel nanoconjugates | Glioblastoma | Apoptosis | [120] |
| tFNA loaded with Temozolomide | Glioblastoma | Apoptosis, Autophagy | [70] |
| tFNA | Parkison’s disease | Apoptosis, differentiation | [135] |
| tFNA | Alzheimer’s disease | Apoptosis | [136] |
| tFNA | Retinal ischemia-reperfusion | Apoptosis | [137] |
| tFNA | Spinal cord injury | Apoptosis | [138] |
| tFNA loaded with SiCCR2 | Intracranial hemorrhage | Anti-inflammation | [139] |
| tFNA | Facial nerve injury | Proliferation, differentiation | [124] |
| tFNA with microRNA-22-3p | Glaucoma | Apoptosis, proliferation | [140] |
| tFNA with Vitamin B12 | Parkinson’s disease | Autophagy, proliferation, differentiation | [141] |
| DNA Nanopore Design | Membrane Anchor | a* | b* | c* | Notable Feature | Ref. |
|---|---|---|---|---|---|---|
| Four-helix bundle | Cholesterol | 0.8 | 11 | 4 | Ion conduction through a lipid bilayer | [149] |
| Six-helix bundle | Cholesterol | 2 | 9 | 3 | Selective transport of small molecules with different charge | [154] |
| Barrel shape | Cholesterol | 2 | 47 | 26 | Transport of DNA hairpin and G-quadruplex | [155] |
| Square funnel shape | Cholesterol | 6 × 6 | 54 | 19 | The first largest synthetic pore | [156] |
| Wireframe cube | Cholesterol | 7 × 7 | 7 | 8 | First open-walled DNA nanopore | [150] |
| Single duplex | Tetraphenylporphyrin | 5 | 6 | Ion-channel made from single DNA duplex | [157] | |
| Six-helix bundle | Tetraphenylporphyrin | 2 | 14 | 2 | Nanopore with two bifunctional tags | [148] |
| Six-helix bundle | Tetraphenylporphyrin | 2 | 14 | 2 | Low conductance occurs at a higher voltage | [158] |
| Six-helix bundle | Alkylphosphorothiolates | 2 | 15 | 72 | Nanopore with modified DNA hydrophobicity | [153] |
| Six-helix bundle 4 × 4 double helix octagon | Alkylphosphorothiolates Cholesterol | 2 35 | 15 10 | 72 32 | Design Simulation Transport of large macromolecules such as folded proteins | [159] [160] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aye, S.L.; Sato, Y. Therapeutic Applications of Programmable DNA Nanostructures. Micromachines 2022, 13, 315. https://doi.org/10.3390/mi13020315
Aye SL, Sato Y. Therapeutic Applications of Programmable DNA Nanostructures. Micromachines. 2022; 13(2):315. https://doi.org/10.3390/mi13020315
Chicago/Turabian StyleAye, Seaim Lwin, and Yusuke Sato. 2022. "Therapeutic Applications of Programmable DNA Nanostructures" Micromachines 13, no. 2: 315. https://doi.org/10.3390/mi13020315