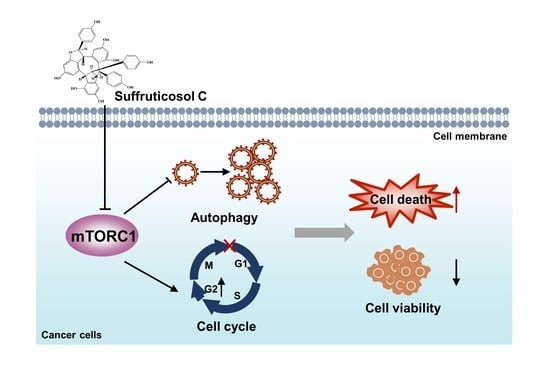
Suffruticosol C-Mediated Autophagy and Cell Cycle Arrest via Inhibition of mTORC1 Signaling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Culture
2.3. Quantitative RT-PCR Analysis
2.4. EdU Assay
2.5. Cell Cycle Analysis
2.6. Ferroptosis Analysis
2.7. Autophagy Analysis
2.8. Cell Apoptosis
2.9. Cell Viability Assay
2.10. Western-Blotting (WB)
2.11. Statistical Analysis
3. Results
3.1. Effect of Suffruticosol C on the Growth of Various Cancer Cells
3.2. Apoptosis-Independent Inhibitory Property of Suffruticosol C on the Growth of Various Cancer Cells
3.3. Ferroptosis-Independent Inhibitory Property of Suffruticosol C on the Growth of Various Cancer Cells
3.4. Autophagy-Dependent Induction of Cell Death by Suffruticosol C
3.5. Induction of Cell Death by Suffruticosol C via Cell Cycle Arrest
3.6. Inhibition of mTORC1 Activation by Suffruticosol C
3.7. Suffruticosol C-Induced Autophagy and Cell Proliferation via Inhibition of the mTORC1 Pathway
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, S.S.; Yuan, R.Y.; Chen, L.G.; Wang, L.S.; Hao, X.H.; Wang, L.J.; Zheng, X.C.; Du, H. Systematic qualitative and quantitative assessment of fatty acids in the seeds of 60 tree peony (Paeonia section Moutan DC.) cultivars by GC-MS. Food Chem. 2015, 173, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.B.; Nam, Y.A.; Kim, H.S.; Hayes, A.W.; Lee, B.M. α-Linolenic acid: Nutraceutical, pharmacological and toxicological evaluation. Food Chem. Toxicol. 2014, 70, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, F.; Ambigaipalan, P. Omega-3 Polyunsaturated Fatty Acids and Their Health Benefits. Annu. Rev. Food. Sci. Technol. 2018, 9, 345–381. [Google Scholar] [CrossRef] [PubMed]
- Valletta, A.; Iozia, L.M.; Leonelli, F. Impact of Environmental Factors on Stilbene Biosynthesis. Plants 2021, 10, 90. [Google Scholar] [CrossRef]
- Snyder, S.A.; Gollner, A.; Chiriac, M.I. Regioselective reactions for programmable resveratrol oligomer synthesis. Nature 2011, 474, 461–466. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Shen, J.; Wang, Z.; Liu, S.; Liu, Q.; Li, Y.; He, C.; Xiao, P.J. Genus Paeonia: A comprehensive review on traditional uses, phytochemistry, pharmacological activities, clinical application, and toxicology. Ethnopharmacol. 2021, 269, 113708. [Google Scholar] [CrossRef]
- Bai, Z.Z.; Tang, J.M.; Ni, J.; Zheng, T.T.; Zhou, Y.; Sun, D.Y.; Li, G.N.; Liu, P.; Niu, L.X.; Zhang, Y.L. Comprehensive metabolite profile of multi-bioactive extract from tree peony (Paeonia ostii and Paeonia rockii) fruits based on MS/MS molecular networking. Food Res. Int. 2021, 148, 110609. [Google Scholar] [CrossRef]
- Liu, Z.; Li, M.; Qian, D.; Liu, Z.; Shu, Q. Phytochemical profiles and the hypoglycemic effects of tree peony seed coats. Food Funct. 2021, 12, 11777–11789. [Google Scholar] [CrossRef]
- Rainer, B.; Revoltella, S.; Mayr, F.; Moesslacher, J.; Scalfari, V.; Kohl, R.; Waltenberger, B.; Pagitz, K.; Siewert, B.; Schwaiger, S.; et al. From bench to counter: Discovery and validation of a peony extract as tyrosinase inhibiting cosmeceutical. Eur. J. Med. Chem. 2019, 184, 111738. [Google Scholar] [CrossRef]
- Liu, P.; Wang, Y.; Gao, J.; Lu, Z.; Yin, W.; Deng, R. Resveratrol trimers from seed cake of Paeonia rockii. Molecules 2014, 19, 19549–19556. [Google Scholar] [CrossRef]
- He, C.N.; Peng, Y.; Xu, L.J.; Liu, Z.A.; Gu, J.; Zhong, A.G.; Xiao, P.G. Three new oligostilbenes from the seeds of Paeonia suffruticosa. Chem. Pharm. Bull. 2010, 58, 843–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almatroodi, S.A.; Alsahli, M.A.; Aljohani, A.S.M.; Alhumaydhi, F.A.; Babiker, A.Y.; Khan, A.A.; Rahmani, A.H. Potential therapeutic targets of resveratrol, a plant polyphenol, and its role in the therapy of various types of cancer. Molecules 2022, 27, 2665. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef] [Green Version]
- Rubinsztein, D.C.; Codogno, P.; Levine, B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug. Discov. 2012, 11, 709–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, K.E.; Losier, T.T.; Russell, R.C. Regulation of Autophagy Enzymes by Nutrient Signaling. Trends Biochem. Sci. 2021, 46, 687–700. [Google Scholar] [CrossRef]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009, 20, 1981–1991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, C.H.; Jun, C.B.; Ro, S.H.; Kim, Y.M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef] [Green Version]
- Ganley, I.G.; du, H.L.; Wang, J.; Ding, X.; Chen, S.; Jiang, X.J. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. Biol. Chem. 2009, 284, 12297–12305. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell. Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [Green Version]
- Settembre, C.; Fraldi, A.; Medina, D.L.; Ballabio, A. Signals from the lysosome: A control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell. Biol. 2013, 14, 283–296. [Google Scholar] [CrossRef]
- Williams, G.H.; Stoeber, K.J. The cell cycle and cancer. Pathology 2012, 226, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, G.K.; Shah, M.A.J. Targeting the cell cycle: A new approach to cancer therapy. Clin. Oncol. 2005, 23, 9408–9421. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Schade, A.E.; Branigan, T.B.; Muller, G.A.; DeCaprio, J.A. Coordinating gene expression during the cell cycle. Trends Biochem. Sci. 2022, 47, 1009–1022. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.Y.; Lim, S.C.; Lee, T.B.; Han, S.I. Molecular Basis of Resveratrol-Induced Resensitization of Acquired Drug-Resistant Cancer Cells. Nutrients 2022, 14, 699. [Google Scholar] [CrossRef]
- Dowling, R.J.; Topisirovic, I.; Alain, T.; Bidinosti, M.; Fonseca, B.D.; Petroulakis, E.; Wang, X.; Larsson, O.; Selvaraj, A.; Liu, Y.; et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 2010, 328, 1172–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-Pozuelo, J.; Figlia, G.; Kaya, O.; Martin-Villalba, A.; Teleman, A.A. Cdk4 and Cdk6 Couple the Cell-Cycle Machinery to Cell Growth via mTORC1. Cell Rep. 2020, 31, 107504. [Google Scholar] [CrossRef] [PubMed]
- Brito, P.M.; Devillard, R.; Negre-Salvayre, A.; Almeida, L.M.; Dinis, T.C.; Salvayre, R.; Auge, N. Resveratrol inhibits the mTOR mitogenic signaling evoked by oxidized LDL in smooth muscle cells. Atherosclerosis 2009, 205, 126–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, R.; Yu, D.; Peng, Y.; Yi, H.; Wang, Y.; Cheng, T.; Shi, B.; Yang, G.; Lai, W.; Wu, X.; et al. Resveratrol induces AMPK and mTOR signaling inhibition-mediated autophagy and apoptosis in multiple myeloma cells. Acta Biochim. Biophys. Sin. 2021, 53, 775–783. [Google Scholar] [CrossRef]
- Liu, M.; Liu, F. Resveratrol inhibits mTOR signaling by targeting DEPTOR. Commun. Integr. Biol. 2011, 4, 382–384. [Google Scholar] [CrossRef] [Green Version]
- Park, D.; Jeong, H.; Lee, M.N.; Koh, A.; Kwon, O.; Yang, Y.R.; Noh, J.; Suh, P.G.; Park, H.; Ryu, S.H. Resveratrol induces autophagy by directly inhibiting mTOR through ATP competition. Sci. Rep. 2016, 6, 21772. [Google Scholar] [CrossRef]
- de Ligt, M.; Bruls, Y.M.H.; Hansen, J.; Habets, M.F.; Havekes, B.; Nascimento, E.B.M.; Moonen-Kornips, E.; Schaart, G.; Schrauwen-Hinderling, V.B.; Lichtenbelt, W.v.; et al. Resveratrol improves ex vivo mitochondrial function but does not affect insulin sensitivity or brown adipose tissue in first degree relatives of patients with type 2 diabetes. Mol. Metab. 2018, 12, 39–47. [Google Scholar] [CrossRef]
- Hu, J.; Lin, T.; Gao, Y.; Xu, J.; Jiang, C.; Wang, G.; Bu, G.; Xu, H.; Chen, H.; Zhang, Y.W. The resveratrol trimer miyabenol C inhibits β-secretase activity and β-amyloid generation. PLoS ONE 2015, 10, e0115973. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Salic, A.; Mitchison, T.J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 2415–2420. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.Y.; Chen, C.W.; Chen, L.K.; Chou, H.Y.; Chang, C.L.; Juan, C.C. Curcumin Attenuates Adipogenesis by Inducing Preadipocyte Apoptosis and Inhibiting Adipocyte Differentiation. Nutrients 2019, 11, 2307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, L.; Jiang, C.; Chen, L.; Jin, J.; Wei, J.; Zhao, L.; Chen, M.; Pan, W.; Xu, Y.; Chu, H.; et al. The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1 activation. Mol. Cell 2015, 58, 804–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Qin, S.; Zheng, Y.; Xia, C.; Zhang, P.; Zhang, L.; Yao, J.; Yi, Y.; Deng, L. T-2 Toxin Induces Ferroptosis by Increasing Lipid Reactive Oxygen Species (ROS) and Downregulating Solute Carrier Family 7 Member 11 (SLC7A11). J. Agric. Food Chem. 2021, 69, 15716–15727. [Google Scholar] [CrossRef] [PubMed]
- Dorrie, J.; Gerauer, H.; Wachter, Y.; Zunino, S.J. Resveratrol induces extensive apoptosis by depolarizing mitochondrial membranes and activating caspase-9 in acute lymphoblastic leukemia cells. Cancer Res. 2001, 61, 4731–4739. [Google Scholar] [PubMed]
- Gao, Y.; He, C.; Ran, R.; Zhang, D.; Li, D.; Xiao, P.G.; Altman, E.J. The resveratrol oligomers, cis- and trans-gnetin H, from Paeonia suffruticosa seeds inhibit the growth of several human cancer cell lines. J. Ethnopharmacol. 2015, 169, 24–33. [Google Scholar] [CrossRef]
- Lee, J.; You, J.H.; Kim, M.S.; Roh, J.L. Epigenetic reprogramming of epithelial-mesenchymal transition promotes ferroptosis of head and neck cancer. Redox Biol. 2020, 37, 101697. [Google Scholar] [CrossRef]
- Li, T.; Tan, Y.; Ouyang, S.; He, J.; Liu, L. Resveratrol protects against myocardial ischemia-reperfusion injury via attenuating ferroptosis. Gene 2022, 808, 145968. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, M.; Qin, C.; Wang, Z.; Chen, J.; Wang, R.; Hu, J.; Zou, Q.; Niu, X. Resveratrol Attenuate Myocardial Injury by Inhibiting Ferroptosis Via Inducing KAT5/GPX4 in Myocardial Infarction. Front. Pharmacol. 2022, 13, 906073. [Google Scholar] [CrossRef] [PubMed]
- Marino, G.; Morselli, E.; Bennetzen, M.V.; Eisenberg, T.; Megalou, E.; Schroeder, S.; Cabrera, S.; Benit, P.; Rustin, P.; Criollo, A.; et al. Longevity-relevant regulation of autophagy at the level of the acetylproteome. Autophagy 2011, 7, 647–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvaraj, S.; Sun, Y.; Sukumaran, P.; Singh, B.B. Resveratrol activates autophagic cell death in prostate cancer cells via downregulation of STIM1 and the mTOR pathway. Mol. Carcinog. 2016, 55, 818–831. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Zepeda, S.P.; Garcia-Villa, E.; Diaz-Chavez, J.; Hernandez-Pando, R.; Gariglio, P. Resveratrol induces cell death in cervical cancer cells through apoptosis and autophagy. Eur. J. Cancer Prev. 2013, 22, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Pozo-Guisado, E.; Alvarez-Barrientos, A.; Mulero-Navarro, S.; Santiago-Josefat, B.; Fernandez-Salguero, P.M. The antiproliferative activity of resveratrol results in apoptosis in MCF-7 but not in MDA-MB-231 human breast cancer cells: Cell-specific alteration of the cell cycle. Biochem. Pharmacol. 2002, 64, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Delmas, D.; Passilly-Degrace, P.; Jannin, B.; Malki, M.C.; Latruffe, N. Resveratrol, a chemopreventive agent, disrupts the cell cycle control of human SW480 colorectal tumor cells. Int. J. Mol. Med. 2002, 10, 193–199. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Sethi, G.; Vadhan-Raj, S.; Bueso-Ramos, C.; Takada, Y.; Gaur, U.; Nair, A.S.; Shishodia, S.; Aggarwal, B.B. Resveratrol inhibits proliferation, induces apoptosis, and overcomes chemoresistance through down-regulation of STAT3 and nuclear factor-kappaB-regulated antiapoptotic and cell survival gene products in human multiple myeloma cells. Blood 2007, 109, 2293–2302. [Google Scholar] [CrossRef] [Green Version]
- Onorati, A.V.; Dyczynski, M.; Ojha, R.; Amaravadi, R.K. Targeting autophagy in cancer. Cancer 2018, 124, 3307–3318. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Yang, G.; Yang, C.; Tang, P.; Chen, J.; Zhang, J.; Liu, J.; Ouyang, L.J. Targeting Autophagy-Related Epigenetic Regulators for Cancer Drug Discovery. Med. Chem. 2021, 64, 11798–11815. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.Q.; Zhang, B.; Wang, L.; Xie, Y.H.; Peng, T.; Bai, B.; Zhou, P.K. Induction of apoptosis and autophagic cell death by the vanillin derivative 6-bromine-5-hydroxy-4-methoxybenzaldehyde is accompanied by the cleavage of DNA-PKcs and rapid destruction of c-Myc oncoprotein in HepG2 cells. Cancer Lett. 2007, 252, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, T.; Kondo, Y.; Kondo, S. Roles of mTOR and STAT3 in autophagy induced by telomere 3’ overhang-specific DNA oligonucleotides. Autophagy 2007, 3, 496–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, J.C.; Shah, M.H.; Ito, T.; Bohas, C.L.; Wolin, E.M.; van Cutsem, E.; Hobday, T.J.; Okusaka, T.; Capdevila, J.; de Vries, E.G.; et al. Everolimus for advanced pancreatic neuroendocrine tumors. N. Engl. J. Med. 2011, 364, 514–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Chen, L.; Qin, S.; Zhang, T.; Yao, J.; Yi, Y.; Deng, L. Mechanistic Target of Rapamycin Complex 1: From a Nutrient Sensor to a Key Regulator of Metabolism and Health. Adv. Nutr. 2022, 13, 1882–1900. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Y.; Sabatini, D.M. Author Correction: mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell. Biol. 2020, 21, 246. [Google Scholar] [CrossRef] [Green Version]
- Martin-Caballero, J.; Flores, J.M.; Garcia-Palencia, P.; Serrano, M. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 2001, 61, 6234–6238. [Google Scholar]
- St Clair, S.; Giono, L.; Varmeh-Ziaie, S.; Resnick-Silverman, L.; Liu, W.J.; Padi, A.; Dastidar, J.; DaCosta, A.; Mattia, M.; Manfredi, J.J. DNA damage-induced downregulation of Cdc25C is mediated by p53 via two independent mechanisms: One involves direct binding to the cdc25C promoter. Mol. Cell. 2004, 16, 725–736. [Google Scholar] [CrossRef]
- Chang, T.C.; Wentzel, E.A.; Kent, O.A.; Ramachandran, K.; Mullendore, M.; Lee, K.H.; Feldmann, G.; Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J.; et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007, 26, 745–752. [Google Scholar] [CrossRef] [Green Version]
- He, L.; He, X.; Lim, L.P.; de Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef]







| Gene | Forward Primer Sequence (5′→3′) | Reverse Primer Sequence (5′→3′) |
|---|---|---|
| CTSA | CAGGCTTTGGTCTTCTCTCCA | TCACGCATTCCAGGTCTTTG |
| TPP1 | GATCCCAGCTCTCCTCAATACG | GCCATTTTTGCACCGTGTG |
| SGSH | TGACCGGCCTTTCTTCCTCTA | GCTCTCTCCGTTGCCAAACTT |
| CSTB | AGTGGAGAATGGCACACCCTA | AAGAAGCCATTGTCACCCCA |
| GLA | AGCCAGATTCCTGCATCAGTG | ATAACCTGCATCCTTCCAGCC |
| SCPEP1 | GATCTCCCCTGTTGATTCGGT | AGCCCCTTATTTACGGCATTC |
| CTSF | ACAGAGGAGGAGTTCCGCACTA | GCTTGCTTCATCTTGTTGCCA |
| ATP6V1H | GGAAGTGTCAGATGATCCCCA | CCGTTTGCCTCGTGGATAAT |
| GAPDH | CAACGAATTTGGCTACAGCA | AGGGGTCTACATGGCAACTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, S.; Geng, H.; Wang, G.; Chen, L.; Xia, C.; Yao, J.; Bai, Z.; Deng, L. Suffruticosol C-Mediated Autophagy and Cell Cycle Arrest via Inhibition of mTORC1 Signaling. Nutrients 2022, 14, 5000. https://doi.org/10.3390/nu14235000
Qin S, Geng H, Wang G, Chen L, Xia C, Yao J, Bai Z, Deng L. Suffruticosol C-Mediated Autophagy and Cell Cycle Arrest via Inhibition of mTORC1 Signaling. Nutrients. 2022; 14(23):5000. https://doi.org/10.3390/nu14235000
Chicago/Turabian StyleQin, Senlin, Huijun Geng, Guoyan Wang, Lei Chen, Chao Xia, Junhu Yao, Zhangzhen Bai, and Lu Deng. 2022. "Suffruticosol C-Mediated Autophagy and Cell Cycle Arrest via Inhibition of mTORC1 Signaling" Nutrients 14, no. 23: 5000. https://doi.org/10.3390/nu14235000