Preparation of Nanoparticle-Loaded Extracellular Vesicles Using Direct Flow Filtration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Quercetin-Iron Complex Nanoparticles
2.3. Cell Culture
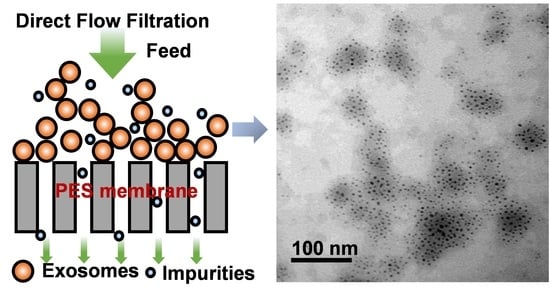
2.4. EV Isolation with Direct Flow Filtration
2.5. SDS-PAGE Gel Electrophoresis
2.6. ExoCheck Antibody Array
2.7. Cellular Uptake and Prussian Blue Staining
2.8. Characterization of Nanoparticles and EVs
3. Results
3.1. Quercetin Iron Complex Nanoparticles
3.2. EV Preparation and Isolation by Direct Flow Filtration
3.3. EV Protein Marker Identification
3.4. Cellular Uptake Studies
4. Discussion
4.1. Quercetin Iron Complex Nanoparticles
4.2. EVs Preparation and Isolation by Direct Flow Filtration
4.3. EV Protein Marker Identification
4.4. Cellular Uptake Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klyachko, N.L.; Arzt, C.J.; Li, S.M.; Gololobova, O.A.; Batrakova, E.V. Extracellular vesicle-based therapeutics: Preclinical and clinical investigations. Pharmaceutics 2020, 12, 1171. [Google Scholar] [CrossRef] [PubMed]
- Yudintceva, N.; Mikhailova, N.; Fedorov, V.; Samochernych, K.; Vinogradova, T.; Muraviov, A.; Shevtsov, M. Mesenchymal stem cells and MSCs-derived extracellular vesicles in infectious diseases: From basic research to clinical practice. Bioengineering 2022, 9, 662. [Google Scholar] [CrossRef] [PubMed]
- Yin, T.; Liu, Y.; Ji, W.; Zhuang, J.; Chen, X.; Gong, B.; Chu, J.; Liang, W.; Gao, J.; Yin, Y. Engineered mesenchymal stem cell-derived extracellular vesicles: A state-of-the-art multifunctional weapon against Alzheimer’s disease. Theranostics 2023, 13, 1264–1285. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef]
- Luan, X.; Sansanaphongpricha, K.; Myers, I.; Chen, H.W.; Yuan, H.B.; Sun, D.X. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017, 38, 754–763. [Google Scholar] [CrossRef]
- Horibe, S.; Tanahashi, T.; Kawauchi, S.; Murakami, Y.; Rikitake, Y. Mechanism of recipient cell-dependent differences in exosome uptake. Bmc Cancer 2018, 18, 47. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Comm. Signaling 2021, 19, 47. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome theranostics: Biology and translational medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef]
- Lobb, R.J.; Lima, L.G.; Moller, A. Exosomes: Key mediators of metastasis and pre-metastatic niche formation. Semin. Cell Dev. Biol. 2017, 67, 3–10. [Google Scholar] [CrossRef]
- Steinbichler, T.B.; Dudas, J.; Riechelmann, H.; Skvortsova, I.I. The role of exosomes in cancer metastasis. Semin. Cancer Biol. 2017, 44, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Elliott, R.O.; He, M. Unlocking the power of exosomes for crossing biological barriers in drug delivery. Pharmaceutics 2021, 13, 122. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Thery, C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J. Extracell. Vesicles 2019, 8, 1648167. [Google Scholar] [CrossRef]
- Margolis, L.; Sadovsky, Y. The biology of extracellular vesicles: The known unknowns. PLoS Biol. 2019, 17, e3000363. [Google Scholar] [CrossRef]
- Singh, K.; Nalabotala, R.; Koo, K.M.; Bose, S.; Nayak, R.; Shiddiky, M.J.A. Separation of distinct exosome subpopulations: Isolation and characterization approaches and their associated challenges. Analyst 2021, 146, 3731–3749. [Google Scholar] [PubMed]
- Nelson, B.C.; Maragh, S.; Ghiran, I.C.; Jones, J.C.; DeRose, P.C.; Elsheikh, E.; Vreeland, W.N.; Wang, L. Measurement and standardization challenges for extracellular vesicle therapeutic delivery vectors. Nanomedicine 2020, 15, 2149–2170. [Google Scholar] [PubMed]
- Bağcı, C.; Sever-Bahcekapili, M.; Belder, N.; Bennett, A.P.S.; Erdener, Ş.E.; Dalkara, T. Overview of extracellular vesicle characterization techniques and introduction to combined reflectance and fluorescence confocal microscopy to distinguish extracellular vesicle subpopulations. Neurophotonics 2022, 9, 021903. [Google Scholar] [CrossRef]
- Perocheau, D.; Touramanidou, L.; Gurung, S.; Gissen, P.; Baruteau, J. Clinical applications for exosomes: Are we there yet? Br. J. Pharmacol. 2021, 178, 2375–2392. [Google Scholar] [CrossRef]
- Amsar, R.M.; Wijaya, C.H.; Ana, I.D.; Hidajah, A.C.; Notobroto, H.B.; Kencana Wungu, T.D.; Barlian, A. Extracellular vesicles: A promising cell-free therapy for cartilage repair. Future Sci. OA 2021, 8, FSO774. [Google Scholar] [CrossRef]
- Wang, X.; Hu, S.; Zhu, D.; Li, J.; Cheng, K.; Liu, G. Comparison of extruded cell nanovesicles and exosomes in their molecular cargos and regenerative potentials. In Nano Research; Springer: Berlin/Heidelberg, Germany, 2023; pp. 1–12. [Google Scholar]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Betzer, O.; Barnoy, E.; Sadan, T.; Elbaz, I.; Braverman, C.; Liu, Z.; Popovtzer, R. Advances in imaging strategies for in vivo tracking of exosomes. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1594. [Google Scholar] [CrossRef]
- Barjesteh, T.; Mansur, S.; Bao, Y.P. Inorganic nanoparticle-loaded exosomes for biomedical applications. Molecules 2021, 26, 1135. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, K.; Liu, Y.; Xu, Y.; Zhang, F.; Yang, H.; Liu, J.; Pan, T.; Chen, J.; Wu, M.; et al. Exosomes mediate the cell-to-cell transmission of IFN-alpha-induced antiviral activity. Nat. Immunol. 2013, 14, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Ni, Z.; Sun, H.; Wang, C. Microfluidic approaches toward the isolation and detection of exosome nanovesicles. Ieee Access 2019, 7, 45080–45098. [Google Scholar] [CrossRef]
- Gardiner, C.; Di Vizio, D.; Sahoo, S.; Thery, C.; Witwer, K.W.; Wauben, M.; Hill, A.F. Techniques used for the isolation and characterization of extracellular vesicles: Results of a worldwide survey. J. Extracell. Vesicles 2016, 5, 32945. [Google Scholar] [CrossRef]
- Schindler, C.; Collinson, A.; Matthews, C.; Pointon, A.; Jenkinson, L.; Minter, R.R.; Vaughan, T.J.; Tigue, N.J. Exosomal delivery of doxorubicin enables rapid cell entry and enhanced in vitro potency. PLoS ONE 2019, 14, 0214545. [Google Scholar] [CrossRef]
- Patel, G.K.; Khan, M.A.; Bhardwaj, A.; Srivastava, S.K.; Zubair, H.; Patton, M.C.; Singh, S.; Khushman, M.; Singh, A.P. Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK. Br. J. Cancer 2017, 116, 609–619. [Google Scholar] [CrossRef]
- Théry, C.; Clayton, A.; Amigorena, S.; Raposo, G.a. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Prot. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef]
- Lai, R.C.; Yeo, R.W.Y.; Tan, K.H.; Lim, S.K. Exosomes for drug delivery—A novel application for the mesenchymal stem cell. Biotechnol. Adv. 2013, 31, 543–551. [Google Scholar] [CrossRef]
- Enderle, D.; Spiel, A.; Coticchia, C.M.; Berghoff, E.; Mueller, R.; Schlumpberger, M.; Sprenger-Haussels, M.; Shaffer, J.M.; Lader, E.; Skog, J.; et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS ONE 2015, 10, e0136133. [Google Scholar] [CrossRef]
- Busatto, S.; Vilanilam, G.; Ticer, T.; Lin, W.L.; Dickson, D.W.; Shapiro, S.; Bergese, P.; Wolfram, J. Tangential flow filtration for highly efficient concentration of extracellular vesicles from large volumes of fluid. Cells 2018, 7, 273. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, T.; Leonardi, A.; Bonaccorso, A.; Pignatello, R.; Puglisi, G. Tangential flow filtration technique: An overview on nanomedicine applications. Pharm. Nanotechnol. 2018, 6, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Park, J.; Jung, J.H.; Lee, R.; Park, J.H.; Yuk, J.M.; Hwang, H.; Yeon, J.H. Cyclic tangential flow filtration system for isolation of extracellular vesicles. Apl Bioeng. 2021, 5, 01613. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, M.; Lucas, K.; Flax, J.; McGrath, J.; Gaborski, T. Tangential flow microfluidics for the capture and release of nanoparticles and extracellular vesicles on conventional and ultrathin membranes. Adv. Mater. Technol. 2019, 4, 1900539. [Google Scholar] [CrossRef] [PubMed]
- Christy, C.; Adams, G.; Kuriyel, R.; Bolton, G.; Seilly, A. High-performance tangential flow filtration: A highly selective membrane separation process. Desalination 2002, 144, 133–136. [Google Scholar] [CrossRef]
- McNamara, R.P.; Caro-Vegas, C.P.; Costantini, L.M.; Landis, J.T.; Griffith, J.D.; Damania, B.A.; Dittmer, D.P. Large-scale, cross-flow based isolation of highly pure and endocytosis-competent extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1541396. [Google Scholar] [CrossRef] [PubMed]
- Imbrogno, A.; Schäfer, A.I. Comparative study of nanofiltration membrane characterization devices of different dimension and configuration (cross flow and dead end). J. Membr. Sci. 2019, 585, 67–80. [Google Scholar] [CrossRef]
- Becht, N.O.; Malik, D.J.; Tarleton, E.S. Evaluation and comparison of protein ultrafiltration test results: Dead-end stirred cell compared with a cross-flow system. Sep. Purif. Technol. 2008, 62, 228–239. [Google Scholar] [CrossRef]
- Fitzsimmons, J.N.; Boyle, E.A. Assessment and comparison of Anopore and cross flow filtration methods for the determination of dissolved iron size fractionation into soluble and colloidal phases in seawater. Limnol. Oceanogr. Methods 2014, 12, 246–263. [Google Scholar] [CrossRef]
- Haraszti, R.A.; Miller, R.; Stoppato, M.; Sere, Y.Y.; Coles, A.; Didiot, M.-C.; Wollacott, R.; Sapp, E.; Dubuke, M.L.; Li, X.; et al. Exosomes produced from 3D cultures of MSCs by tangential flow filtration show higher yield and improved activity. Mol. Ther. J. Am. Soc. Gene Ther. 2018, 26, 2838–2847. [Google Scholar] [CrossRef]
- Gonzalez-Gonzalez, L.; Gonzalez-Ramirez, R.; Flores, A.; Avelino-Cruz, J.E.; Felix, R.; Monjaraz, E. Epidermal growth factor potentiates migration of MDA-MB 231 breast cancer cells by increasing Na(V)1.5 channel expression. Oncology 2019, 97, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Aktas, C.C.; Zeybek, N.D.; Piskin, A.K. In vitro effects of phenytoin and DAPT on MDA-MB-231 breast cancer cells. Acta Biochim. Biophys. Sin. 2015, 47, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Chamaraux-Tran, T.N.; Mathelin, C.; Aprahamian, M.; Joshi, G.P.; Tomasetto, C.; Diemunsch, P.; Akladios, C. Antitumor effects of lidocaine on human breast cancer cells: An in vitro and in vivo experimental trial. Anticancer Res. 2018, 38, 95–105. [Google Scholar] [PubMed]
- Han, J.; Lim, W.; You, D.; Jeong, Y.; Kim, S.; Lee, J.E.; Shin, T.H.; Lee, G.; Park, S. Chemoresistance in the human triple-negative breast cancer cell line MDA-MB-231 induced by doxorubicin gradient is associated with epigenetic alterations in histone deacetylase. J. Oncol. 2019, 2019, 1345026. [Google Scholar] [CrossRef]
- Papan, P.; Kantapan, J.; Sangthong, P.; Meepowpan, P.; Dechsupa, N. Iron (III)-quercetin complex: Synthesis, physicochemical characterization, and MRI cell tracking toward potential applications in regenerative medicine. Contrast Media Mol. Imaging 2020, 2020, 8877862. [Google Scholar] [CrossRef]
- Massi, A.; Bortolini, O.; Ragno, D.; Bernardi, T.; Sacchetti, G.; Tacchini, M.; De Risi, C. Research progress in the modification of quercetin leading to anticancer agents. Molecules 2017, 22, 1270. [Google Scholar] [CrossRef]
- Xu, D.; Hu, M.; Wang, Y.; Cui, Y. Antioxidant activities of quercetin and its complexes for medicinal application. Molecules 2019, 24, 1123. [Google Scholar] [CrossRef] [PubMed]
- Leopoldini, M.; Russo, N.; Chiodo, S.; Toscano, M. Iron chelation by the powerful antioxidant flavonoid quercetin. J. Agr. Food Chem. 2006, 54, 6343–6351. [Google Scholar] [CrossRef]
- da Silva, W.M.B.; de Oliveira Pinheiro, S.; Alves, D.R.; de Menezes, J.; Magalhães, F.E.A.; Silva, F.C.O.; Silva, J.; Marinho, E.S.; de Morais, S.M. Synthesis of quercetin-metal complexes, in vitro and in silico anticholinesterase and antioxidant evaluation, and in vivo toxicological and anxiolitic activities. Neurotox. Res. 2020, 37, 893–903. [Google Scholar] [CrossRef]
- Raza, A.; Xu, X.; Xia, L.; Xia, C.; Tang, J.; Ouyang, Z. Quercetin-iron complex: Synthesis, characterization, antioxidant, DNA binding, DNA cleavage, and antibacterial activity studies. J. Fluoresc. 2016, 26, 2023–2031. [Google Scholar] [CrossRef]
- Faria, A.; Meireles, M.; Fernandes, I.; Santos-Buelga, C.; Gonzalez-Manzano, S.; Duenas, M.; de Freitas, V.; Mateus, N.; Calhau, C. Flavonoid metabolites transport across a human BBB model. Food Chem. 2014, 149, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, R.G.R.; Pinheiro, M.; Neves, A.R. Nanotechnology innovations to enhance the therapeutic efficacy of quercetin. Nanomaterials 2021, 11, n2658. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, J.; Rich, M.; Lovas, K.; Warram, J.; Bolding, M.S.; Bao, Y. T-1-Enhanced MRI-visible nanoclusters for imaging-guided drug delivery. Nanoscale 2017, 9, 11785–11792. [Google Scholar] [CrossRef]
- Prestianni, L.; Espinal, E.R.; Hathcock, S.F.; Vollmuth, N.; Wang, P.; Holler, R.A.; Liu, S.; Kim, B.J.; Bao, Y. Synthesis and Characterization of quercetin–iron complex nanoparticles for overcoming drug resistance. Pharmaceutics 2023, 15, 1041. [Google Scholar] [CrossRef] [PubMed]
- Narkhede, A.A.; Sherwood, J.A.; Antone, A.; Coogan, K.R.; Bolding, M.S.; Deb, S.; Bao, Y.P.; Rao, S.S. Role of surface chemistry in mediating the uptake of ultrasmall iron oxide nanoparticles by cancer cells. Acs Appl. Mater. Interfaces 2019, 11, 17157–17166. [Google Scholar] [CrossRef]
- Xu, Y.; Sherwood, J.A.; Lackey, K.H.; Qin, Y.; Bao, Y. The responses of immune cells to iron oxide nanoparticles. J. Appl. Toxicol. 2016, 36, 543–553. [Google Scholar] [CrossRef]
- Sherwood, J.; Lovas, K.; Rich, M.; Yin, Q.; Lackey, K.; Bolding, M.S.; Bao, Y. Shape-dependent cellular behaviors and relaxivity of iron oxide-based T-1 MRI contrast agents. Nanoscale 2016, 8, 17506–17515. [Google Scholar] [CrossRef]
- Kim, J.Y.; Rhim, W.K.; Yoo, Y.I.; Kim, D.; Ko, K.W.; Heo, Y.; Park, C.G.; Han, D.K. Defined MSC exosome with high yield and purity to improve regenerative activity. J. Tissue Eng. 2021, 12, 20417314211008626. [Google Scholar] [CrossRef]
- Salmond, N.; Williams, K.C. Isolation and characterization of extracellular vesicles for clinical applications in cancer—Time for standardization? Nanoscale Adv. 2021, 3, 1830–1852. [Google Scholar] [CrossRef]
- Clavijo Rivera, E.; Villafaña-López, L.; Liu, S.; Vinoth Kumar, R.; Viau, M.; Bourseau, P.; Monteux, C.; Frappart, M.; Couallier, E. Cross-flow filtration for the recovery of lipids from microalgae aqueous extracts: Membrane selection and performances. Process Biochem. 2020, 89, 199–207. [Google Scholar] [CrossRef]
- Mocé-Llivina, L.; Jofre, J.; Muniesa, M. Comparison of polyvinylidene fluoride and polyether sulfone membranes in filtering viral suspensions. J. Virol. Methods 2003, 109, 99–101. [Google Scholar] [CrossRef] [PubMed]
- Kadel, S.; Pellerin, G.; Thibodeau, J.; Perreault, V.; Lainé, C.; Bazinet, L. How molecular weight cut-offs and physicochemical properties of polyether sulfone membranes affect peptide migration and selectivity during electrodialysis with filtration membranes. Membranes 2019, 9, 153. [Google Scholar] [CrossRef] [PubMed]
- Vergauwen, G.; Dhondt, B.; Van Deun, J.; De Smedt, E.; Berx, G.; Timmerman, E.; Gevaert, K.; Miinalainen, I.; Cocquyt, V.; Braems, G.; et al. Confounding factors of ultrafiltration and protein analysis in extracellular vesicle research. Sci. Rep. 2017, 7, 2704. [Google Scholar] [CrossRef]
- Abello, J.; Nguyen, T.; Marasini, R.; Weiss, M.; Aryal, S. Biodistribution of gadolinium-and near infrared-labeled human umbilical cord mesenchymal stromal cell-derived exosomes in tumor bearing mice. Theranostics 2019, 9, 2325–2345. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Sancho-Albero, M.; Encabo-Berzosa, M.D.; Beltran-Visiedo, M.; Fernandez-Messina, L.; Sebastian, V.; Sanchez-Madrid, F.; Arruebo, M.; Santamaria, J.; Martin-Duque, P. Efficient encapsulation of theranostic nanoparticles in cell-derived exosomes: Leveraging the exosomal biogenesis pathway to obtain hollow gold nanoparticle-hybrids. Nanoscale 2019, 11, 18825–18836. [Google Scholar] [PubMed]
- Pan, S.; Zhao, X.; Shao, C.; Fu, B.; Huang, Y.; Zhang, N.; Dou, X.; Zhang, Z.; Qiu, Y.; Wang, R.; et al. STIM1 promotes angiogenesis by reducing exosomal miR-145 in breast cancer MDA-MB-231 cells. Cell Death Dis. 2021, 12, 38. [Google Scholar] [CrossRef]
- González-King, H.; Tejedor, S.; Ciria, M.; Gil-Barrachina, M.; Soriano-Navarro, M.; Sánchez-Sánchez, R.; Sepúlveda, P.; García, N.A. Non-classical Notch signaling by MDA-MB-231 breast cancer cell-derived small extracellular vesicles promotes malignancy in poorly invasive MCF-7 cells. Cancer Gene Ther. 2022, 29, 1056–1069. [Google Scholar] [CrossRef]
- Pols, M.S.; Klumperman, J. Trafficking and function of the tetraspanin CD63. Exp. Cell. Res. 2009, 315, 1584–1592. [Google Scholar] [CrossRef]
- Kreger, B.T.; Johansen, E.R.; Cerione, R.A.; Antonyak, M.A. The enrichment of survivin in exosomes from breast cancer cells treated with paclitaxel promotes cell survival and chemoresistance. Cancers 2016, 8, 111. [Google Scholar] [CrossRef]
- Chiba, M.; Kimura, M.; Asari, S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol. Rep. 2012, 28, 1551–1558. [Google Scholar] [CrossRef] [PubMed]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.I.E.; Lehtiö, J.; El Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, D.K.; Hvam, M.L.; Primdahl-Bengtson, B.; Boysen, A.T.; Whitehead, B.; Dyrskjøt, L.; Ørntoft, T.F.; Howard, K.A.; Ostenfeld, M.S. Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J. Extracell. Vesicles 2014, 3, 25011. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.D.; Patnam, S.; Koyyada, R.; Samal, R.; Alvi, S.B.; Satyanaryana, G.; Andrews, R.; Panigrahi, A.K.; Rengan, A.K.; Mudigonda, S.S.; et al. Identifying stable reference genes in polyethene glycol precipitated urinary extracellular vesicles for RT-qPCR-based gene expression studies in renal graft dysfunction patients. Transplant Immunol. 2022, 75, 101715. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Sancho-Albero, M.; Navascues, N.; Mendoza, G.; Sebastian, V.; Arruebo, M.; Martin-Duque, P.; Santamaria, J. Exosome origin determines cell targeting and the transfer of therapeutic nanoparticles towards target cells. J. Nanobiotechnol. 2019, 17, 16. [Google Scholar] [CrossRef]
- Fu, S.; Wang, Y.; Xia, X.; Zheng, J.C. Exosome engineering: Current progress in cargo loading and targeted delivery. NanoImpact 2020, 20, 100261. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mansur, S.; Habib, S.; Hawkins, M.; Brown, S.R.; Weinman, S.T.; Bao, Y. Preparation of Nanoparticle-Loaded Extracellular Vesicles Using Direct Flow Filtration. Pharmaceutics 2023, 15, 1551. https://doi.org/10.3390/pharmaceutics15051551
Mansur S, Habib S, Hawkins M, Brown SR, Weinman ST, Bao Y. Preparation of Nanoparticle-Loaded Extracellular Vesicles Using Direct Flow Filtration. Pharmaceutics. 2023; 15(5):1551. https://doi.org/10.3390/pharmaceutics15051551
Chicago/Turabian StyleMansur, Shomit, Shahriar Habib, Mikayla Hawkins, Spenser R. Brown, Steven T. Weinman, and Yuping Bao. 2023. "Preparation of Nanoparticle-Loaded Extracellular Vesicles Using Direct Flow Filtration" Pharmaceutics 15, no. 5: 1551. https://doi.org/10.3390/pharmaceutics15051551