Carriers of ADAMTS13 Rare Variants Are at High Risk of Life-Threatening COVID-19
Abstract
:1. Introduction
2. Materials and Methods
2.1. GEN-COVID Cohort
2.2. Whole Exome Sequencing Analysis (WES)
2.3. Phenotype Definition Adjusting by Age
2.4. ADAMTS13 Assay
2.5. Statistical Analysis
3. Results
3.1. ADAMTS13 Ultra-Rare Variants Associate with Severity in COVID-19
3.2. Characterization of Ultra-Rare Variants
3.3. Characterization of ADAMTS13 Activity of Ultra-Rare Variants
3.4. Laboratory Values in Heterozygous Subjects
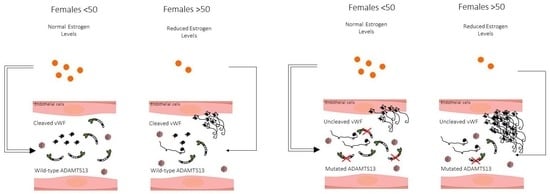
3.5. Autosomal Dominant Disorder Conditioned by SARS-CoV-2 Infection, Sex and Age
3.6. Pediatric Cases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pagliari, M.T.; Cairo, A.; Boscarino, M.; Mancini, I.; Pappalardo, E.; Bucciarelli, P.; Martinelli, I.; Rosendaal, F.R.; Peyvandi, F. Role of ADAMTS13, VWF and F8 genes in deep vein thrombosis. PLoS ONE 2021, 16, e0258675. [Google Scholar] [CrossRef]
- Haybar, H.; Khodadi, E.; Kavianpour, M.; Saki, N. Mutations and Common Polymorphisms in ADAMTS13 and vWF Genes Responsible for Increasing Risk of Thrombosis. Cardiovasc. Hematol. Disord. Drug Targets 2018, 18, 176–181. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Tang, N.; Li, D.; Wang, X.; Sun, Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020, 18, 1233–1234. [Google Scholar] [CrossRef] [Green Version]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Qu, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Cao, B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Hafez, W.; Ziade, M.A.; Arya, A.; Saleh, H.; Ali, S.; Rao, S.R.; Alla, O.F.; Ali, M.; Al Zouhbi, M.; Abdelrahman, A. Reduced ADAMTS13 Activity in Correlation with Pathophysiology, Severity, and Outcome of COVID-19: A Retrospective Observational Study. Int. J. Infect. Dis. 2022, 117, 334–344. [Google Scholar] [CrossRef]
- Powazniak, Y.; Kempfer, A.C.; Pereyra, J.C.; Palomino, J.P.; Lazzari, M.A. VWF and ADAMTS13 behavior in estradiol-treated HUVEC. Eur. J. Haematol. 2011, 86, 140–147. [Google Scholar] [CrossRef]
- Fallerini, C.; Picchiotti, N.; Baldassarri, M.; Zguro, K.; Daga, S.; Fava, F.; Benetti, E.; Amitrano, S.; Bruttini, M.; Palmieri, M.; et al. Common, low-frequency, rare, and ultra-rare coding variants contribute to COVID-19 severity. Hum. Genet. 2022, 141, 147–173. [Google Scholar] [CrossRef]
- Daga, S.; Fallerini, C.; Baldassarri, M.; Fava, F.; Valentino, F.; Doddato, G.; Benetti, E.; Furini, S.; Giliberti, A. Employing a systematic approach to biobanking and analyzing clinical and genetic data for advancing COVID-19 research. Eur. J. Hum. Genet. 2021, 29, 745–759. [Google Scholar] [CrossRef]
- Picchiotti, N.; Benetti, E.; Fallerini, C.; Daga, S.; Baldassarri, M.; Fava, F.; Zguro, K.; Valentino, F.; Doddato, G.; Giliberti, A.; et al. Post-Mendelian genetic model in COVID-19. Cardiol. Cardiovasc. Med. 2021, 5, 673–694. [Google Scholar] [CrossRef]
- WHO. COVID-19 Therapeutic Trial Synopsis; WHO R & D Blueprint Novel Coronavirus; COVID-19 Therapeutic Trial Synopsis; R & D Blueprint; WHO: New York, NY, USA, 2020. [Google Scholar]
- Chiasakul, T.; Cuker, A. Clinical and laboratory diagnosis of TTP: An integrated approach. Hematol. Am. Soc. Hematol. Educ. Program. 2018, 2018, 530–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, H.M. Pathophysiology of thrombotic thrombocytopenic purpura. Int. J. Hematol. 2010, 91, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Scully, M.; Cataland, S.; Coppo, P.; De La Rubia, J.; Friedman, K.D.; Hovinga, J.A.K.; Lämmle, B.; Matsumoto, M.; Pavenski, K.; Sadler, E.; et al. Consensus on the standardization of terminology in thrombotic thrombocytopenic purpura and related thrombotic microangiopathies. J. Thromb. Haemost. 2017, 15, 312–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, B.; Kaur, P.; Mekheal, E.; Fasulo, S.; Maroules, M. COVID-19 and thrombotic thrombocytopenic purpura: A review of literature. Hematol. Transfus. Cell Ther. 2021, 43, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Kremer Hovinga, J.A.; George, J.N. Hereditary Thrombotic Thrombocytopenic Purpura. N. Engl. J. Med. 2019, 381, 1653–1662. [Google Scholar] [CrossRef] [PubMed]
- Alwan, F.; Vendramin, C.; Liesner, R.; Clark, A.; Lester, W.; Dutt, T.; Thomas, W.; Gooding, R.; Biss, T.; Watson, H.G.; et al. Characterization and treatment of congenital thrombotic thrombocytopenic purpura. Blood 2019, 133, 1644–1651. [Google Scholar] [CrossRef] [Green Version]
- Page, E.E.; Kremer Hovinga, J.A.; Terrell, D.R.; Vesely, S.K.; George, J.N. Thrombotic thrombocytopenic purpura: Diagnostic criteria, clinical features, and long-term outcomes from 1995 through 2015. Blood Adv. 2017, 1, 590–600. [Google Scholar] [CrossRef]
- Laffan, M. Can you grow out of von Willebrand disease? Haemoph. Off. J. World Fed. Hemoph. 2017, 23, 807–809. [Google Scholar] [CrossRef] [Green Version]
- Ward, S.E.; O’Sullivan, J.M.; O’Donnell, J.S. The relationship between ABO blood group, von Willebrand factor, and primary hemostasis. Blood 2020, 136, 2864–2874. [Google Scholar] [CrossRef]
- Graviilaki, E.; Kokoris, S.; Touloumenidouet, T.; Koravou, E.; Koutra, M.; Karali, V.; Anagnostopoulos, A. Thrombotic microangiopathy variants are independently associated with critical disease in COVID-19 patients. Blood 2020, 136 (Suppl. S1), 21–22. [Google Scholar]
- Panigada, M.; Bottino, N.; Tagliabue, P.; Grasselli, G.; Novembrino, C.; Chantarangkul, V.; Pesenti, A.; Peyvandi, F.; Tripodi, A. Hypercoagulability of COVID-19 patients in intensive care unit: A report of thromboelastography findings and other parameters of hemostasis. J. Thromb. Haemost. 2020, 18, 1738–1742. [Google Scholar] [CrossRef] [PubMed]
- Joly, B.S.; Darmon, M.; Dekimpe, C. Imbalance of von Willebrand factor and ADAMTS13 axis is rather a biomarker of strong inflammation and endothelial damage than a cause of thrombotic process in critically ill COVID-19 patients. J. Thromb. Haemost. 2021, 19, 2193–2198. [Google Scholar] [CrossRef] [PubMed]
- Pascreau, T.; Zia-Chahabi, S.; Zuber, B.; Tcherakian, C.; Farfour, E.; Vasse, M. ADAMTS 13 deficiency is associated with abnormal distribution of von Willebrand factor multimers in patients with COVID-19. Thromb. Res. 2021, 204, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, G.; Maji, M.; Mandal, S.; Vaniyath, S.; Negi, G.; Nath, U.K. COVID 19 infection associated with thrombotic thrombocytopenic purpura. J. Thromb. Thrombolysis 2021, 52, 504–507. [Google Scholar] [CrossRef]
- Mancini, I.; Baronciani, L.; Artoni, A.; Colpani, P.; Biganzoli, M.; Cozzi, G.; Peyvandi, F. The ADAMTS13-von Willebrand factor axis in COVID-19 patients. J. Thromb. Haemost. 2021, 19, 513–521. [Google Scholar] [CrossRef]

 ). The inheritance pattern appears that of an autosomal dominant disorder conditioned by SARS-CoV-2 infection, sex, and age.
). The inheritance pattern appears that of an autosomal dominant disorder conditioned by SARS-CoV-2 infection, sex, and age.
 ). The inheritance pattern appears that of an autosomal dominant disorder conditioned by SARS-CoV-2 infection, sex, and age.
). The inheritance pattern appears that of an autosomal dominant disorder conditioned by SARS-CoV-2 infection, sex, and age.
| (a) | |||
| Phenotype | Ultra-Rare Variants | Wild Type | Total |
| Severe | 32 | 454 | 486 |
| Not severe | 6 | 283 | 289 |
| Total | 38 | 737 | 775 (Grand Total) |
| OR = 3.32 (95% CI 1.37 to 8.05); p-value = 4.9 × 10−3 | |||
| (b) | |||
| Phenotype | Ultra-Rare Variants | Wild Type | Total |
| Severe | 23 | 649 | 672 |
| Not severe | 26 | 526 | 552 |
| Total | 49 | 1175 | 1224 (Grand Total) |
| p-value = 0.252888 | |||
| Nucleotide Change | Amino Acid Change | dbSNP | CADD | ExAC_NFE | Tot. n. Patients | Sex (n.) | Age Range | Hospitalized (Not Hospitalized) | Category † |
|---|---|---|---|---|---|---|---|---|---|
| c.11G > A | p.R4H | rs370406676 | 5.2 | 0.0001 | 1 | F | 56 | 1 | 2 |
| c.220C > T | p.R74W | n.a. | 22 | 0.000008 | 1 | M | 39 | 1 | 1 |
| c.241C > T | p.H81Y | rs148644959 | 23 | n.a | 4 | M (2) | 38–60 | 2 | 1 |
| F (2) | 60–68 | 2 | 1 | ||||||
| c.353C > T | p.P118L | rs587698109 | 19.3 | 0.000008 | 1 | F | 59 | 1 | 3 |
| c.559G > C | p.D187H | rs148312697 | 25.6 | 0.0006 | 7 | M(5) | 41–76 | 4 | 3–2 |
| 54 | (1) | 0 | |||||||
| F (2) | 50–82 | 2 | 3–2 | ||||||
| c.649G > A | p.D217N | rs782305581 | 29.4 | 0.00003 | 2 | M(2) | 75 | 1 | 3 |
| 34 | (1) | 0 | |||||||
| c.703G > T | p.D235Y | n.a. | 33 | 0.00004 | 1 | F | 45 | 1 | 2 |
| c.722G > C | p.G241A | n.a. | 8.1 | n.a. | v1 | F | 30 | (1) | 0 |
| c.742G > A | p.V248M | n.a. | 25.1 | 0.00004 | 2 | M | 49–51 | 2 | 3–2 |
| c.953A > G | p.K318R | n.a. | 0.006 | n.a. | 1 | F | 42 | (1) | 0 |
| c.1016C > G | p.T339R | rs149517360 | 22.8 | 0.0004 # | 6 | M | 40 | 1 | 3 |
| F (5) | 9 months-66 | 4 | 2–1 | ||||||
| 23 | (1) | 0 | |||||||
| c.1084G > A | p.V362M | rs781924046 | 0.21 | 0.00001523 | 1 | F | 46 | (1) | 0 |
| c.1117_1121del | p.S373Gfs*15 | n.a | n.a | n.a | 1 | M | 39 | 1 | 1 |
| c.1157G > A | p.R386H | rs151048660 | 11.6 | 0.0003 | 10 | F (5) | 46–82 | 4 | 4–3 |
| 49 | (1) | 0 | |||||||
| M (4) | 42–73 | 4 | 3–2 | ||||||
| c.1178G > A | p.R393Q | rs140937290 | 12.5 | 0.000017 | 1 | M | 68 | 1 | 1 |
| c.1226G > A | p.R409Q | n.a | 35 | n.a. | 1 | M | 48 | 1 | 1 |
| c.1261C > T | p.R421C | rs145825553 | 33 | 0.0008 | 6 | M (6) | 55–81 | 4 | 3–1 |
| 52–65 | (2) | 0 | |||||||
| c.1291G > A | p.E431Q | rs781915989 | 25.8 | 0.000018 | 2 | M | 47 | (1) | 0 |
| F | 75 | (1) | 0 | ||||||
| c.1336A > G | p.M446V | rs782733359 | 16.1 | 0.000022 | 1 | F | 70 | 1 | 3 |
| c.1423C > T | p.P475S | rs11575933 | 4.5 | 0.0006 § | 3 | M (2) | 33 | 1 | 1 |
| 63 | (1) | 0 | |||||||
| F | 1 month | 1 | 1 | ||||||
| c.1463G > A | p.R488Q | rs147201977 | 22.6 | n.a. | 1 | M | 72 | 1 | 5 |
| c.1486A > G | p.M496V | rs782574335 | 0.001 | 0.00004 | 1 | F | 44 | 1 | 1 |
| c.1492C > A | p.R498S | n.a. | 32 | n.a. | 1 | F | 93 | 1 | 2 |
| c.1601G > A | p.G534D | rs782003053 | 26.6 | 0.00005 | 1 | M | 62 | 1 | 2 |
| c.1700C > T | p.A567V | rs782272645 | 27.4 | 0.00007 | 1 | M | 79 | 1 | 2 |
| c.1729A > T | p.T577S | n.a. | 8.6 | n.a. | 1 | F | 33 | 1 | 1 |
| c.1753A > G | p.I585V | n.a. | 0.001 | n.a. | 1 | M | 82 | 1 | 3 |
| c.1808A > G | p.Y603C | rs867154790 | 24.2 | n.a. | 1 | M | 52 | 1 | 1 |
| c.1906C > T | p.R636W | rs201704847 | 24.7 | 0.000008 | 2 | M | 57–62 | 2 | 3–2 |
| c.1931G > A | p.R644H | rs782184721 | 0.011 | 0.00002 | 1 | F | 70 | 1 | 2 |
| c.1976G > A | p.R659K | rs150764227 | 23.5 | 0.0003 | 1 | M | 57 | 1 | 2 |
| c.2009G > A | p.R670H | rs149953167 | 10.7 | 0.0003 | 1 | F | 78 | 1 | 5 |
| c.2011C > A | p.P671T | n.a. | 22.1 | n.a. | 1 | F | 56 | 1 | 3 |
| c.2038C > T | p.P680S | n.a. | 24.3 | n.a. | 1 | M | 76 | 1 | 2 |
| c.2099G > A | p.G700E | n.a. | 31 | n.a. | 1 | M | 25 | 1 | 3 |
| c.2111G > A | p.R704H | rs782223605 | 23.7 | 0.0000008 | 1 | F | 34 | 1 | 3 |
| c.2111G > T | p.R704L | n.a. | 26.2 | n.a. | 1 | M | 68 | 1 | 2 |
| c.2278G > A | p.G760S | rs782729939 | 22.8 | 0.00005 | 1 | F | 57 | 1 | 2 |
| c.2282G > T | p.G761V | n.a. | 25.3 | n.a. | 1 | F | 49 | (1) | 0 |
| c.2288G > A | p.R763Q | rs781804540 | 16.8 | 0.000020 | 1 | M | 60 | 1 | 2 |
| c.2351G > A | p.R784Q | rs377187626 | 4.4 | n.a. | 1 | M | 57 | 1 | 2 |
| c.2420G > C | p.R807T | n.a. | 23.5 | n.a. | 1 | M | 71 | 1 | 2 |
| c.2422C > T | p.W808R | n.a. | 0.007 | n.a. | 1 | M | 50 | (1) | 0 |
| c.2494G > A | p.V832M | rs34104386 | 18.5 | 0.000017 ^ | 2 | M | 28 | 1 | 1 |
| F | 7 months | 1 | 2 | ||||||
| c.2519C > T | p.A840V | n.a. | 0.3 | n.a. | 1 | F | 67 | 1 | 2 |
| c.2545G > A | p.V849I | rs140639242 | 0.4 | 0.0002 | 1 | M | 72 | 1 | 5 |
| c.2773A > G | p.R925G | rs782263547 | 4.1 | 0.000009 | 2 | M | 57–65 | 2 | 4–3 |
| c.2814G > T | p.K938N | n.a. | 25.7 | n.a. | 2 | M | 57–72 | 2 | 4–2 |
| c.2824C > T | p.R942W | rs929435102 | 27.7 | 0.000009 | 2 | M | 61 | (1) | 0 |
| F | 56 | 1 | 2 | ||||||
| c.2828G > A | p.R943Q | rs782160285 | 2.6 | 0.00009 | 1 | M | 84 | 1 | 5 |
| c.2854C > T | p.P952S | rs143568784 | 29.9 | 0.0003 | 5 | M | 68 | 1 | 2 |
| F (4) | 67–85 | 3 | 2 | ||||||
| 40 | (1) | 0 | |||||||
| c.2915G > A | p.R972Q | rs139951127 | 5.4 | 0.0002 | 7 | M (4) | 37–78 | 3 | 3–2 |
| 50 | 1 | 0 | |||||||
| F (3) | 35–70 | 1 | 5–1 | ||||||
| c.2978C > T | p.T993I | rs139808736 | 23.2 | 0.00006 | 1 | M | 57 | 1 | 3 |
| c.3161delC | p.Cys1055Valfs*66 | n.a. | n.a. | n.a. | 1 | F | 54 | 1 | 2 |
| c.3201T > A | p.C1067 * | n.a. | 36 | n.a. | 1 | M | 72 | 1 | 3 |
| c.3356C > T | p.P1119L | rs1044262941 | 36 | 0.000009 | 1 | M | 64 | 1 | 2 |
| c.3463G > A | p.A1155T | n.a. | 1.6 | n.a | 1 | M | 37 | 1 | 2 |
| c.3541G > A | p.G1181R | rs192619276 | 1.5 * | 0.000009 ° | 5 | M (3) | 34–74 | 3 | 3–1 |
| F (2) | 57–66 | 2 | 3–2 | ||||||
| c.3685G > A | p.V1229I | rs587643681 | 2.5 | 0.00001769 | 1 | M | 56 | 1 | 1 |
| c.3694A > T | p.S1232C | n.a. | 23.6 | 0.00001769 | 1 | M | 50 | 1 | 1 |
| c.3713C > T | p.A1238V | rs587697598 | 13.9 | 0.00007986 | 1 | F | 63 | 1 | 3 |
| c.3718G T | p.D1240Y | n.a. | 24.9 | n.a. | 1 | M | 60 | 1 | 2 |
| c.3722T > C | p.M1241T | rs1057522240 | 0.002 | 0.000008 | 1 | F | 46 | 1 | 1 |
| c.3740G > A | p.R1247Q | rs782197792 | 27.2 | 0.00004 | 1 | M | 34 | (1) | 0 |
| c.3826G > A | p.G1276R | rs144808448 | 0.493 | 0.00003 | 1 | M | 62 | 1 | 5 |
| c.3853C > T | p.R1285W | rs370157837 | 27.6 | 0.00002264 | 1 | M | 68 | 1 | 4 |
| c.3956C > T | p.T1319M | rs375824927 | 8.19 | n.a. | 1 | F | 84 | 1 | 2 |
| c.3962A > T | p.N1321I | rs200645384 | 1.248 | 0.00006 | 1 | M | 73 | 1 | 2 |
| c.4007G > A | p.R1336Q | rs782213090 | 23.8 | 0.000008 | 1 | M | 53 | 1 | 2 |
| c.4012G > A | p.A1338T | rs782401854 | 27 | 0.000008 | 1 | M | 60 | 1 | 3 |
| c.4141T > G | p.S1381A | n.a. | 25.6 | n.a. | 1 | F | 29 | (1) | 0 |
| c.4262_4271del | p.G1423Efs*6 | n.a. | n.a. | n.a. | 1 | M | 48 | 1 | 3 |
| CRP M and F cases | CRP M < 50 y cases | CRP F ≥ 50 y cases | ||||||
| Ultra-rare variants | Mean | Count | Ultra-rare variants | Mean | Count | Ultra-rare variants | Mean | Count |
| yes | 39 | 64 | yes | 24.5 | 12 | yes | 55.1 | 21 |
| no | 28.5 | 1491 | no | 18.7 | 163 | no | 27.1 | 443 |
| p-value = 0.0005166 | p-value = 0.1116 | p-value = 0.0001896 | ||||||
| Fibrinogen M and F cases | Fibrinogen M < 50 y cases | Fibrinogen F ≥ 50 y cases | ||||||
| Ultra-rare variants | Mean | Count | Ultra-rare variants | Mean | Count | Ultra-rare variants | Mean | Count |
| yes | 488 | 25 | yes | 446 | 6 | yes | 502 | 8 |
| no | 502 | 806 | no | 499 | 67 | no | 503 | 234 |
| p-value = 0.8843 | p-value = 0.6227 | p-value = 0.9406 | ||||||
| D-Dimer M and F cases | D-Dimer M < 50 y cases | D-Dimer F ≥ 50 y cases | ||||||
| Ultra-rare variants | Mean | Count | Ultra-rare variants | Mean | Count | Ultra-rare variants | Mean | Count |
| yes | 3024 | 58 | yes | 4016 | 8 | yes | 4127 | 18 |
| no | 2788 | 1446 | no | 1908 | 148 | no | 2711 | 441 |
| p-value = 0.03431 | p-value = 0.1889 | p-value = 0.47 | ||||||
| Platelets M and F cases | Platelets M <50 y cases | Platelets F ≥ 50 y cases | ||||||
| Ultra-rare variants | Mean | Count | Ultra-rare variants | Mean | Count | Ultra-rare variants | Mean | Count |
| yes | 183 | 63 | yes | 153 | 11 | yes | 182 | 17 |
| no | 315 | 1509 | no | 221 | 184 | no | 574 | 451 |
| p-value = 0.07816 | p-value = 0.05286 | p-value = 0.2213 | ||||||
| LDH M and F cases | LDH M <50 y cases | LDH F ≥ 50 y cases | ||||||
| Ultra-rare variants | Mean | Count | Ultra-rare variants | Mean | Count | Ultra-rare variants | Mean | Count |
| yes | 444 | 49 | yes | 513 | 8 | yes | 506 | 16 |
| no | 395 | 1313 | no | 392 | 142 | no | 374 | 389 |
| p-value = 0.009494 | p-value = 0.05761 | p-value = 0.006933 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zguro, K.; Baldassarri, M.; Fava, F.; Beligni, G.; Daga, S.; Leoncini, R.; Galasso, L.; Cirianni, M.; Rusconi, S.; Siano, M.; et al. Carriers of ADAMTS13 Rare Variants Are at High Risk of Life-Threatening COVID-19. Viruses 2022, 14, 1185. https://doi.org/10.3390/v14061185
Zguro K, Baldassarri M, Fava F, Beligni G, Daga S, Leoncini R, Galasso L, Cirianni M, Rusconi S, Siano M, et al. Carriers of ADAMTS13 Rare Variants Are at High Risk of Life-Threatening COVID-19. Viruses. 2022; 14(6):1185. https://doi.org/10.3390/v14061185
Chicago/Turabian StyleZguro, Kristina, Margherita Baldassarri, Francesca Fava, Giada Beligni, Sergio Daga, Roberto Leoncini, Lucrezia Galasso, Michele Cirianni, Stefano Rusconi, Matteo Siano, and et al. 2022. "Carriers of ADAMTS13 Rare Variants Are at High Risk of Life-Threatening COVID-19" Viruses 14, no. 6: 1185. https://doi.org/10.3390/v14061185