In Vitro Diagnostic Assay to Detect SARS-CoV-2-Neutralizing Antibody in Patient Sera Using Engineered ACE-2 Mini-Protein
Abstract
:1. Introduction
2. Material and Methods
2.1. Protein Expression and Purification
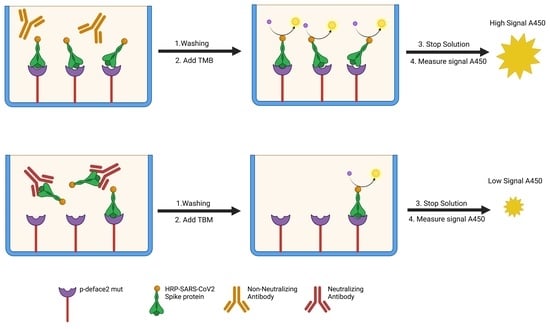
2.2. SARS-CoV-2-Neutralizing Antibody ELISA
2.3. Calculation and Interpretation of the Results
2.4. Detection of SARS-CoV-2 Spike Protein Variants
2.5. Cell Toxicity Analysis
2.6. Neutralization Assay
3. Results
3.1. SARS-CoV-2-Neutralizing Antibody ELISA
3.2. Neutralization Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Weekly Epidemiological Update on COVID-19-21 September 2022. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---21-september-2022 (accessed on 21 November 2022).
- Moghadas, S.M.; Vilches, T.N.; Zhang, K.; Wells, C.R.; Shoukat, A.; Singer, B.H.; Meyers, L.A.; Neuzil, K.M.; Langley, J.M.; Fitzpatrick, M.C.; et al. The Impact of Vaccination on COVID-19 Outbreaks in the United States. medRxiv 2021, 73, 2257–2264. [Google Scholar] [CrossRef]
- Coronavirus Disease 2019. Available online: https://www.cdc.gov/media/releases/2021/p0607-mrna-reduce-risks.html (accessed on 21 November 2022).
- Johnson, A.G. COVID-19 Incidence and Death Rates Among Unvaccinated and Fully Vaccinated Adults with and without Booster Doses During Periods of Delta and Omicron Variant Emergence—25 U.S. Jurisdictions, April 4–December 25 2021. MMWR Morb. Mortal. Wkly. Rep. 2022, 71. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, F.; Shen, C.; Peng, W.; Li, D.; Zhao, C.; Li, Z.; Li, S.; Bi, Y.; Yang, Y.; et al. A Noncompeting Pair of Human Neutralizing Antibodies Block COVID-19 Virus Binding to Its Receptor ACE2. Science 2020, 368, 1274–1278. [Google Scholar] [CrossRef]
- Assadiasl, S.; Fatahi, Y.; Zavvar, M.; Nicknam, M.H. COVID-19: Significance of Antibodies. Hum. Antibodies 2020, 28, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Arvin, A.M.; Fink, K.; Schmid, M.A.; Cathcart, A.; Spreafico, R.; Havenar-Daughton, C.; Lanzavecchia, A.; Corti, D.; Virgin, H.W. A Perspective on Potential Antibody-Dependent Enhancement of SARS-CoV-2. Nature 2020, 584, 353–363. [Google Scholar] [CrossRef] [PubMed]
- The Germinal Centre B Cell Response to SARS-CoV-2|Nature Reviews Immunology. Available online: https://www.nature.com/articles/s41577-021-00657-1 (accessed on 21 November 2022).
- Martínez-Flores, D.; Zepeda-Cervantes, J.; Cruz-Reséndiz, A.; Aguirre-Sampieri, S.; Sampieri, A.; Vaca, L. SARS-CoV-2 Vaccines Based on the Spike Glycoprotein and Implications of New Viral Variants. Front. Immunol. 2021, 12, 701501. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Kyriakidis, N.C.; López-Cortés, A.; González, E.V.; Grimaldos, A.B.; Prado, E.O. SARS-CoV-2 Vaccines Strategies: A Comprehensive Review of Phase 3 Candidates. npj Vaccines 2021, 6, 28. [Google Scholar] [CrossRef]
- Krammer, F. SARS-CoV-2 Vaccines in Development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- Murin, C.D.; Wilson, I.A.; Ward, A.B. Antibody Responses to Viral Infections: A Structural Perspective across Three Different Enveloped Viruses. Nat. Microbiol. 2019, 4, 734–747. [Google Scholar] [CrossRef]
- Pang, N.Y.-L.; Pang, A.S.-R.; Chow, V.T.; Wang, D.-Y. Understanding Neutralising Antibodies against SARS-CoV-2 and Their Implications in Clinical Practice. Mil. Med. Res. 2021, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, J.; Xu, X.; Liao, G.; Chen, Y.; Hu, C.-H. Patterns of IgG and IgM Antibody Response in COVID-19 Patients. Emerg. Microbes Infect. 2020, 9, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Valdez-Cruz, N.A.; García-Hernández, E.; Espitia, C.; Cobos-Marín, L.; Altamirano, C.; Bando-Campos, C.G.; Cofas-Vargas, L.F.; Coronado-Aceves, E.W.; González-Hernández, R.A.; Hernández-Peralta, P.; et al. Integrative Overview of Antibodies against SARS-CoV-2 and Their Possible Applications in COVID-19 Prophylaxis and Treatment. Microb. Cell Factories 2021, 20, 88. [Google Scholar] [CrossRef] [PubMed]
- Seow, J.; Graham, C.; Merrick, B.; Acors, S.; Pickering, S.; Steel, K.J.A.; Hemmings, O.; O’Byrne, A.; Kouphou, N.; Galao, R.P.; et al. Longitudinal Observation and Decline of Neutralizing Antibody Responses in the Three Months Following SARS-CoV-2 Infection in Humans. Nat. Microbiol. 2020, 5, 1598–1607. [Google Scholar] [CrossRef]
- Marot, S.; Malet, I.; Leducq, V.; Zafilaza, K.; Sterlin, D.; Planas, D.; Gothland, A.; Jary, A.; Dorgham, K.; Bruel, T.; et al. Rapid Decline of Neutralizing Antibodies against SARS-CoV-2 among Infected Healthcare Workers. Nat. Commun. 2021, 12, 844. [Google Scholar] [CrossRef]
- Sterlin, D.; Mathian, A.; Miyara, M.; Mohr, A.; Anna, F.; Claër, L.; Quentric, P.; Fadlallah, J.; Devilliers, H.; Ghillani, P.; et al. IgA Dominates the Early Neutralizing Antibody Response to SARS-CoV-2. Sci. Transl. Med. 2021, 13, eabd2223. [Google Scholar] [CrossRef]
- Figueiredo-Campos, P.; Blankenhaus, B.; Mota, C.; Gomes, A.; Serrano, M.; Ariotti, S.; Costa, C.; Nunes-Cabaço, H.; Mendes, A.M.; Gaspar, P.; et al. Seroprevalence of Anti-SARS-CoV-2 Antibodies in COVID-19 Patients and Healthy Volunteers up to 6 Months Post Disease Onset. Eur. J. Immunol. 2020, 50, 2025–2040. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody Responses to SARS-CoV-2 in Patients with Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2027–2034. [Google Scholar] [CrossRef]
- Zhang, W.; Du, R.-H.; Li, B.; Zheng, X.-S.; Yang, X.-L.; Hu, B.; Wang, Y.-Y.; Xiao, G.-F.; Yan, B.; Shi, Z.-L.; et al. Molecular and Serological Investigation of 2019-NCoV Infected Patients: Implication of Multiple Shedding Routes. Emerg. Microbes Infect. 2020, 9, 386–389. [Google Scholar] [CrossRef] [Green Version]
- Cervia, C.; Nilsson, J.; Zurbuchen, Y.; Valaperti, A.; Schreiner, J.; Wolfensberger, A.; Raeber, M.E.; Adamo, S.; Weigang, S.; Emmenegger, M.; et al. Systemic and Mucosal Antibody Responses Specific to SARS-CoV-2 during Mild versus Severe COVID-19. J. Allergy Clin. Immunol. 2021, 147, 545–557.e9. [Google Scholar] [CrossRef]
- Robust Neutralizing Antibodies to SARS-CoV-2 Infection Persist for Months|Science. Available online: https://www.science.org/doi/10.1126/science.abd7728 (accessed on 21 November 2022).
- Alsoussi, W.B.; Malladi, S.K.; Zhou, J.Q.; Liu, Z.; Ying, B.; Kim, W.; Schmitz, A.J.; Lei, T.; Horvath, S.C.; Sturtz, A.J.; et al. SARS-CoV-2 Omicron Boosting Induces de Novo B Cell Response in Humans. bioRxiv 2022. [Google Scholar] [CrossRef]
- Piechotta, V.; Harder, T. Waning of COVID-19 Vaccine Effectiveness: Individual and Public Health Risk. Lancet 2022, 399, 887–889. [Google Scholar] [CrossRef] [PubMed]
- Feikin, D.R.; Higdon, M.M.; Abu-Raddad, L.J.; Andrews, N.; Araos, R.; Goldberg, Y.; Groome, M.J.; Huppert, A.; O’Brien, K.L.; Smith, P.G.; et al. Duration of Effectiveness of Vaccines against SARS-CoV-2 Infection and COVID-19 Disease: Results of a Systematic Review and Meta-Regression. Lancet 2022, 399, 924–944. [Google Scholar] [CrossRef] [PubMed]
- Shete, A.M.; Patil, D.Y.; Sahay, R.R.; Sapkal, G.N.; Deshpande, G.R.; Yadav, P.D. Waning Natural and Vaccine-Induced Immunity Leading to Reinfection with SARS-CoV-2 Omicron Variant. Hum. Vaccin. Immunother. 2022, 2127289. [Google Scholar] [CrossRef]
- Turner, J.S.; O’Halloran, J.A.; Kalaidina, E.; Kim, W.; Schmitz, A.J.; Zhou, J.Q.; Lei, T.; Thapa, M.; Chen, R.E.; Case, J.B.; et al. SARS-CoV-2 MRNA Vaccines Induce Persistent Human Germinal Centre Responses. Nature 2021, 596, 109–113. [Google Scholar] [CrossRef]
- Amanat, F.; Thapa, M.; Lei, T.; Ahmed, S.M.S.; Adelsberg, D.C.; Carreño, J.M.; Strohmeier, S.; Schmitz, A.J.; Zafar, S.; Zhou, J.Q.; et al. SARS-CoV-2 MRNA Vaccination Induces Functionally Diverse Antibodies to NTD, RBD, and S2. Cell 2021, 184, 3936–3948.e10. [Google Scholar] [CrossRef]
- Sønderskov, K.M.; Vistisen, H.T.; Dinesen, P.T.; Østergaard, S.D. COVID-19 Booster Vaccine Willingness. Dan Med. J. 2021, 69, A10210765. [Google Scholar]
- Klugar, M.; Riad, A.; Mohanan, L.; Pokorná, A. COVID-19 Vaccine Booster Hesitancy (VBH) of Healthcare Workers in Czechia: National Cross-Sectional Study. Vaccines 2021, 9, 1437. [Google Scholar] [CrossRef]
- Troiano, G.; Nardi, A. Vaccine Hesitancy in the Era of COVID-19. Public Health 2021, 194, 245–251. [Google Scholar] [CrossRef]
- Fernandes, L.A.; Gomes, A.A.; Guimarães, B.G.; de Lourdes Borba Magalhães, M.; Ray, P.; da Silva, G.F. Engineering Defensin α-Helix to Produce High-Affinity SARS-CoV-2 Spike Protein Binding Ligands. Protein Sci. 2022, 31, e4355. [Google Scholar] [CrossRef]
- Carlin, A.F.; Clark, A.E.; Chaillon, A.; Garretson, A.F.; Bray, W.; Porrachia, M.; Santos, A.T.; Rana, T.M.; Smith, D.M. Virologic and Immunologic Characterization of COVID-19 Recrudescence after Nirmatrelvir/Ritonavir Treatment. Clin. Infect. Dis. 2022, ciac496. [Google Scholar] [CrossRef] [PubMed]
- DeWolf, S.; Laracy, J.C.; Perales, M.-A.; Kamboj, M.; van den Brink, M.R.M.; Vardhana, S. SARS-CoV-2 in Immunocompromised Individuals. Immunity 2022, 55, 1779–1798. [Google Scholar] [CrossRef] [PubMed]




| Bioclin Assay 1 | Mini Protein Assay 2 | ||||||
|---|---|---|---|---|---|---|---|
| Sample | Result | Abs (450 nm) | Neutralization (%) | Abs (450 nm) | Abs (450 nm) | Abs Mean | Neutralization (%) |
| 1 | Negative | 2.141 | 5.47 | 0.535 | 0.509 | 0.522 | 0 |
| 2 | Negative | 2.178 | 3.84 | 0.427 | 0.409 | 0.418 | 19.9 |
| 3 | Negative | 1.901 | 16.07 | 0.506 | 0.324 | 0.415 | 20.4 |
| 4 | Negative | 1.892 | 5.40 | 0.45 | 0.30 | 0.375 | 28.1 |
| 5 | Negative | 1.950 | 6.91 | 0.465 | 0.412 | 0.439 | 15.9 |
| 6 | Negative | 2.093 | 7.59 | 0.38 | 0.269 | 0.325 | 37.8 |
| 7 | Negative | 2.450 | 8.16 | 0.462 | 0.376 | 0.419 | 19.7 |
| 8 | Negative | 2.086 | 14.36 | 0.448 | 0.41 | 0.429 | 17.8 |
| 9 | Negative | 2.412 | 6.49 | 0.396 | 0.477 | 0.437 | 16.3 |
| 10 | Negative | 1.859 | 7.05 | 0.365 | 0.316 | 0.341 | 34.7 |
| 11 | Negative | 1.949 | 13.95 | 0.507 | 0.425 | 0.466 | 10.7 |
| 12 | Negative | 2.342 | 3.40 | 0.349 | 0.335 | 0.342 | 34.4 |
| 13 | Positive | 0.894 | 55.65 | 0.233 | 0.186 | 0.210 | 59.8 |
| 14 | Positive | 1.114 | 41.37 | 0.339 | 0.296 | 0.318 | 39.1 |
| 15 | Positive | 0.912 | 54.76 | 0.245 | 0.182 | 0.214 | 59.0 |
| 16 | Positive | 0.617 | 66.17 | 0.277 | 0.26 | 0.269 | 48.5 |
| 17 | Positive | 0.745 | 63.05 | 0.266 | 0.258 | 0.262 | 49.8 |
| 18 | Positive | 0.956 | 47.59 | 0.193 | 0.173 | 0.183 | 64.9 |
| 19 | Positive | 0.725 | 64.04 | 0.27 | 0.287 | 0.279 | 46.6 |
| 20 | Positive | 0.106 | 94.74 | 0.153 | 0.165 | 0.159 | 69.5 |
| 21 | Positive | 0.224 | 88.21 | 0.18 | 0.217 | 0.199 | 61.9 |
| Number of total tested samples | 21 |
| Number of truly negative samples | 12 |
| Number of truly positive samples | 9 |
| Number of false-positives | 0 |
| Number of false-negative | 0 |
| Cut-off | 38% |
| Sensitivity | 100% |
| Specificity | 100% |
| NT50 (µg/mL) | SEM | |
|---|---|---|
| h-deface2 | 8.9 × 104 | 4.7 × 105 |
| p-deface2 | 1.2 × 103 | 1.5 × 103 |
| p-deface-mut | 3.8 × 102 | 2.6 × 102 |
| p-deface-ala | 3.1 × 102 | 1.9 × 102 |
| anti-spike | 9.1 × 10−2 | 8.8 × 10−3 |
| no treatment | 1.1 × 105 | 2.0 × 106 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira de Jesus, B.A.; Gomes, A.A.; Clark, A.E.; Rodrigues, T.A.; Ledgerwood-Lee, M.; Van Zant, W.; Brickner, H.; Wang, M.; Blum, D.L.; Cassera, M.B.; et al. In Vitro Diagnostic Assay to Detect SARS-CoV-2-Neutralizing Antibody in Patient Sera Using Engineered ACE-2 Mini-Protein. Viruses 2022, 14, 2823. https://doi.org/10.3390/v14122823
Pereira de Jesus BA, Gomes AA, Clark AE, Rodrigues TA, Ledgerwood-Lee M, Van Zant W, Brickner H, Wang M, Blum DL, Cassera MB, et al. In Vitro Diagnostic Assay to Detect SARS-CoV-2-Neutralizing Antibody in Patient Sera Using Engineered ACE-2 Mini-Protein. Viruses. 2022; 14(12):2823. https://doi.org/10.3390/v14122823
Chicago/Turabian StylePereira de Jesus, Bruna Andersen, Anderson Albino Gomes, Alex E. Clark, Tayse Andrade Rodrigues, Melissa Ledgerwood-Lee, Westley Van Zant, Howard Brickner, Meiqiao Wang, David L. Blum, Maria B. Cassera, and et al. 2022. "In Vitro Diagnostic Assay to Detect SARS-CoV-2-Neutralizing Antibody in Patient Sera Using Engineered ACE-2 Mini-Protein" Viruses 14, no. 12: 2823. https://doi.org/10.3390/v14122823