A Novel RNA Virus, Macrobrachium rosenbergii Golda Virus (MrGV), Linked to Mass Mortalities of the Larval Giant Freshwater Prawn in Bangladesh
Abstract
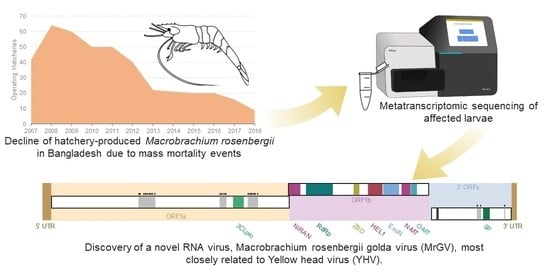
:1. Introduction
2. Materials and Methods
2.1. Collection of Hatchery Cultured Larvae and Wild Broodstock
2.2. Preparation of Infectious Study Material
2.3. In Vivo Tank Experiments
- Experimental larvae were directly immersed into filtered challenge medium from moribund larvae for 10 min prior to being transferred to a fibreglass tank.
- Experimental larvae were fed artemia that had been immersed in challenge medium from moribund larvae for 10 min.
- Experimental larvae were immersed into filtered challenge medium from healthy larvae for 10 min prior to being transferred to a fibreglass tank.
2.4. RNA Extraction
2.5. Double Stranded cDNA Synthesis
2.6. Library Preparation
2.7. Sequence Analysis
2.8. Phylogenetic Analyses
2.9. Primer Design and Specific PCR
2.10. Specific PCR Screens
3. Results
3.1. In Vivo Tank Experiments
3.2. Complete Genome Assembly of Virus
3.3. Open Reading Frame (ORF) Prediction, Genome Prediction and Motif Identification
3.4. Identification of Frameshift Motifs
3.5. Prediction of 3′ UTR Secondary Structure
3.6. Phylogenetic Characterisation of MrGV
3.7. Multiple Sequence Alignment (MSA) of 3′ ORFs
3.8. Infection Prevalence in Hatchery-Reared Larvae and Wild River-Based Adult Populations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- FAO. FAO Yearbook; FAO Fisheries and Aquaculture Department: Rome, Italy, 2019. [Google Scholar]
- New, M.B.; Nair, C.M. Global scale of freshwater prawn farming. Aquac. Res. 2012, 43, 960–969. [Google Scholar] [CrossRef]
- Pillai, D.; Bonami, J.R. A review on the diseases of freshwater prawns with special focus on white tail disease of Macrobrachium rosenbergii. Aquac. Res. 2012, 43, 1029–1037. [Google Scholar] [CrossRef]
- Wahab, M.A.; Ahmad-Al-Nahid, S.; Ahmed, N.; Haque, M.M.; Karim, M. Current status and prospects of farming the giant river prawn Macrobrachium rosenbergii (De Man) in Bangladesh. Aquac. Res. 2012, 43, 970–983. [Google Scholar] [CrossRef]
- Currie, D.; Basak, S.; SAFETI, Winrock Bangladesh, Dhaka, Bangladesh; Debnath, P.P.; WorldFish Bangladesh, Dhaka, Bangladesh; Islam, H.M.R.; BFRI, Shrimp Research Station, Bagerhat, Bangladesh. Personal communication, 2019.
- Alam, S.; Rahman-Al-Mamun, A.; Hossain, M.T.; Yeasin, M.; Hossain, M. Mass larval mortality in a giant freshwater prawn Macrobrachium rosenbergii hatchery: An attempt to detect microbes in the berried and larvae. Bangladesh J. Fish. 2019, 31, 41–48. [Google Scholar]
- Ahmed, N.; Troell, M. Fishing for prawn larvae in Bangladesh: An important coastal livelihood causing negative effects on the environment. Ambio 2010, 39, 20–29. [Google Scholar] [CrossRef] [Green Version]
- Srisala, J.; Pukmee, R.; McIntosh, R.; Choosuk, S.; Itsathitphaisarn, O.; Flegel, T.W.; Sritunyalucksana, K.; Vanichviriyakit, R. Distinctive histopathology of Spiroplasma eriocheiris infection in the giant river prawn Macrobrachium rosenbergii. Aquaculture 2018, 493, 93–99. [Google Scholar] [CrossRef]
- Arcier, J.M.; Herman, F.; Lightner, D.V.; Redman, R.M.; Mari, J.; Bonami, J.R. A viral disease associated with mortalities in hatchery-reared postlarvae of the giant freshwater prawn Macrobrachium rosenbergii. Dis. Aquat. Organ. 1999, 38, 177–181. [Google Scholar] [CrossRef] [Green Version]
- Qian, D.; Shi, Z.; Zhang, S.; Cao, Z.; Liu, W.; Li, L.; Xie, Y.; Cambournac, I.; Bonami, J.-R. Extra small virus-like particles (XSV) and nodavirus associated with whitish muscle disease in the giant freshwater prawn, Macrobrachium rosenbergii. J. Fish Dis. 2003, 26, 521–527. [Google Scholar] [CrossRef]
- Yoganandhan, K.; Leartvibhas, M.; Sriwongpuk, S. White tail disease of the giant freshwater prawn Macrobrachium rosenbergii in Thailand. Dis. Aquat. Organ. 2006, 69, 255–258. [Google Scholar] [CrossRef] [Green Version]
- Hameed, A.S.S.; Yoganandhan, K.; Widada, J.S. Experimental transmission and tissue tropism of Macrobrachium rosenbergii nodavirus (MrNV) and its associated extra small virus (XSV). Dis. Aquat. Organ. 2004, 62, 191–196. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.S.; Chang, J.S.; Shih, H.H.; Chen, S.N. RT-PCR amplification and sequence analysis of extra small virus associated with white tail disease of Macrobrachium rosenbergii (de Man) cultured in Taiwan. J. Fish Dis. 2007, 30, 127–132. [Google Scholar] [CrossRef]
- Murwantoko, M.; Bimantara, A.; Roosmanto, R.; Kawaichi, M. Macrobrachium rosenbergii nodavirus infection in a giant freshwater prawn hatchery in Indonesia. Springerplus 2016, 5, 1729. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.; Cao, Z.; Yuan, J.; Shi, Z.; Yuan, X.; Lin, L.; Xu, Y.; Yao, J.; Hao, G.; Shen, J. Isolation and Characterization of a Novel Dicistrovirus Associated with Moralities of the Great Freshwater Prawn, Macrobrachium rosenbergii. Int. J. Mol. Sci. 2016, 17, 204. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, C.Y.; Chuang, P.C.; Chen, L.C.; Tu, C.; Chien, M.S.; Huang, K.C.; Kao, H.F.; Tung, M.C.; Tsai, S.S. Infectious hypodermal and haematopoietic necrosis virus (IHHNV) infections in giant freshwater prawn, Macrobrachium rosenbergii. Aquaculture 2006, 258, 73–79. [Google Scholar] [CrossRef]
- Qiu, L.; Chen, X.; Zhao, R.-H.; Li, C.; Gao, W.; Zhang, Q.-L.; Huang, J. Description of a Natural Infection with Decapod Iridescent Virus 1 in Farmed Giant Freshwater Prawn, Macrobrachium rosenbergii. Viruses 2019, 11, 354. [Google Scholar] [CrossRef] [Green Version]
- Gangnonngiw, W.; Kiatpathomchai, W.; Sriurairatana, S.; Laisutisan, K.; Chuchird, N.; Limsuwan, C. Parvo-like virus in the hepatopancreas of freshwater prawns Macrobrachium rosenbergii cultivated in Thailand. Dis. Aquat. Organ. 2009, 85, 167–173. [Google Scholar] [CrossRef] [Green Version]
- Sahul Hameed, A.; Charles, M.X.; Anilkumar, M. Tolerance of Macrobrachium rosenbergii to white spot syndrome virus. Aquaculture 2000, 183, 207–213. [Google Scholar] [CrossRef]
- Sarathi, M.; Nazeer Basha, A.; Ravi, M.; Venkatesan, C.; Senthil Kumar, B.; Sahul Hameed, A.S. Clearance of white spot syndrome virus (WSSV) and immunological changes in experimentally WSSV-injected Macrobrachium rosenbergii. Fish Shellfish Immunol. 2008, 25, 222–230. [Google Scholar] [CrossRef]
- Gangnonngiw, W.; Bunnontae, M.; Phiwsaiya, K.; Senapin, S.; Dhar, A.K. In experimental challenge with infectious clones of Macrobrachium rosenbergii nodavirus (MrNV) and extra small virus (XSV), MrNV alone can cause mortality in freshwater prawn (Macrobrachium rosenbergii). Virology 2020, 540, 30–37. [Google Scholar] [CrossRef]
- Khuntia, C.P.; Das, B.K.; Samantaray, B.R.; Samal, S.K.; Mishra, B.K. Characterization and pathogenicity studies of Vibrio parahaemolyticus isolated from diseased freshwater prawn, Macrobrachium rosenbergii (de Man). Aquac. Res. 2008, 39, 301–310. [Google Scholar] [CrossRef]
- Krueger, F. A Wrapper Tool around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files. Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 30 June 2019).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 30 June 2019).
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Genomes and Mini-metagenomes from Highly Chimeric Reads. In Research in Computational Molecular Biology; Deng, M., Jiang, R., Sun, F., Zhang, X., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 158–170. [Google Scholar] [CrossRef]
- Hunt, M.; Gall, A.; Ong, S.H.; Brener, J.; Ferns, B.; Goulder, P.; Nastouli, E.; Keane, J.A.; Kellam, P.; Otto, T.D. IVA: Accurate de novo assembly of RNA virus genomes. Bioinformatics 2015, 31, 2374–2376. [Google Scholar] [CrossRef] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.-J.; Tappu, R. MEGAN Community Edition-Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. Subgroup, 1000 Genome Project Data Processing The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [Green Version]
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L.M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T.F.; Conesa, A. Qualimap: Evaluating next-generation sequencing alignment data. Bioinformatics 2012, 28, 2678–2679. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.-Y.; Gao, Y.-Z.; Du, M.-Z.; Liu, S.; Dong, C.; Guo, F.-B. Vgas: A Viral Genome Annotation System. Front. Microbiol. 2019, 10, 184. [Google Scholar] [CrossRef]
- Hildebrand, A.; Remmert, M.; Biegert, A.; Söding, J. Fast and accurate automatic structure prediction with HHpred. Proteins Struct. Funct. Bioinform. 2009, 77, 128–132. [Google Scholar] [CrossRef] [Green Version]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Byun, Y.; Moon, S.; Han, K. A general computational model for predicting ribosomal frameshifts in genome sequences. Comput. Biol. Med. 2007, 37, 1796–1801. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Saberi, A.; Gulyaeva, A.A.; Brubacher, J.L.; Newmark, P.A.; Gorbalenya, A.E. A planarian nidovirus expands the limits of RNA genome size. PLoS Pathog. 2018, 14, e1007314. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sahul Hameed, A.S.; Yoganandhan, K.; Sri Widada, J.; Bonami, J.R. Studies on the occurrence of Macrobrachium rosenbergii nodavirus and extra small virus-like particles associated with white tail disease of M. rosenbergii in India by RT-PCR detection. Aquaculture 2004, 238, 127–133. [Google Scholar] [CrossRef]
- Sudhakaran, R.; Yoganandhan, K.; Ahmed, V.P.I. Artemia as a possible vector for Macrobrachium rosenbergii nodavirus (MrNV) and extra small virus transmission (XSV) to Macrobrachium rosenbergii post-larvae. Dis. Aquat. Organ. 2006, 70, 161–166. [Google Scholar] [CrossRef]
- Lo, C.; Leu, J.; Ho, C.; Chen, C.; Peng, S.; Chen, Y.; Chou, C.; Yeh, P.; Huang, C.; Chou, H.; et al. Detection of baculovirus associated with white spot syndrome (WSBV) in penaeid shrimps using polymerase chain reaction. Dis. Aquat. Organ. 1996, 25, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Surachetpong, W.; Poulos, B.T.; Tang, K.F.J.; Lightner, D.V. Improvement of PCR method for the detection of monodon baculovirus (MBV) in penaeid shrimp. Aquaculture 2005, 249, 69–75. [Google Scholar] [CrossRef]
- Ding, Z.; Bi, K.; Wu, T.; Gu, W.; Wang, W.; Chen, J. A simple PCR method for the detection of pathogenic spiroplasmas in crustaceans and environmental samples. Aquaculture 2007, 265, 49–54. [Google Scholar] [CrossRef]
- Mohr, P.; Moody, N.; Hoad, J.; Williams, L.; Bowater, R.; Cummins, D.; Cowley, J.; Crane, M.S. The gene encoding the nucleocapsid protein of Gill-associated nidovirus of Penaeus monodon prawns is located upstream. Dis. Aquat. Organ. 2015, 115, 263–268. [Google Scholar] [CrossRef]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J.B.T. Virus Taxonomy—Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2012; ISBN 9780123846846. [Google Scholar]
- Wijegoonawardane, P.K.M.; Cowley, J.A.; Phan, T.; Hodgson, R.A.J.; Nielsen, L.; Kiatpathomchai, W.; Walker, P.J. Genetic diversity in the yellow head nidovirus complex. Virology 2008, 380, 213–225. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Cowley, J.A.; Cadogan, L.C.; Spann, K.M.; Sittidilokratna, N.; Walker, P.J. The gene encoding the nucleocapsid protein of Gill-associated nidovirus of Penaeus monodon prawns is located upstream of the glycoprotein gene. J. Virol. 2004, 78, 8935–8941. [Google Scholar] [CrossRef] [Green Version]
- Boonyaratpalin, S.; Supamattaya, K.; Kasornchandra, J.; Direkbusaracom, S.; Aekpanithanpong, U.; Chantanachookin, C. Non-occluded baculolike virus, the causative agent of yellow-head disease in the black tiger shrimp (Penaeus monodon). Fish Pathol. 1993, 28, 103–109. [Google Scholar] [CrossRef] [Green Version]
- Lightner, D.V.; Redman, R.M. A baculovirus-caused disease of the penaeid shrimp, Penaeus monodon. J. Invertebr. Pathol. 1981, 38, 299–302. [Google Scholar] [CrossRef]
- Spann, K.M.; Vickers, J.E.; Lester, R.J.G. Lymphoid organ virus of Penaeus monodon from Australia. Dis. Aquat. Organ. 1995, 23, 127–134. [Google Scholar] [CrossRef]
- Limsuwan, C. Handbook for Cultivation of Black Tiger Prawns; Tamsetakit Co., Ltd.: Bangkok, Thailand, 1991. [Google Scholar]
- Fields, B.N.; Knipe, D.M.; Howley, P.M. Fields Virology, 5th ed.; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; ISBN 9780781760607. [Google Scholar]
- Buck, K.W. Comparison of The Replication of Positive-Stranded RNA Viruses of Plants and Animals. Adv. Virus Res. 1996, 47, 159–251. [Google Scholar] [CrossRef]
- Dreher, T.W. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 1999, 37, 151–174. [Google Scholar] [CrossRef]
- Munang’andu, H.M.; Mugimba, K.K.; Byarugaba, D.K.; Mutoloki, S.; Evensen, Ø. Current advances on virus discovery and diagnostic role of viral metagenomics in aquatic organisms. Front. Microbiol. 2017, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Hu, T.; Liu, Q.; Li, C.; Sun, Y.; Wang, Y.; Shi, W.; Zhao, Q.; Huang, J. A Novel Hepe-Like Virus from Farmed Giant Freshwater Prawn Macrobrachium rosenbergii. Viruses 2020, 12, 3234. [Google Scholar] [CrossRef] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hooper, C.; Debnath, P.P.; Biswas, S.; van Aerle, R.; Bateman, K.S.; Basak, S.K.; Rahman, M.M.; Mohan, C.V.; Islam, H.M.R.; Ross, S.; et al. A Novel RNA Virus, Macrobrachium rosenbergii Golda Virus (MrGV), Linked to Mass Mortalities of the Larval Giant Freshwater Prawn in Bangladesh. Viruses 2020, 12, 1120. https://doi.org/10.3390/v12101120
Hooper C, Debnath PP, Biswas S, van Aerle R, Bateman KS, Basak SK, Rahman MM, Mohan CV, Islam HMR, Ross S, et al. A Novel RNA Virus, Macrobrachium rosenbergii Golda Virus (MrGV), Linked to Mass Mortalities of the Larval Giant Freshwater Prawn in Bangladesh. Viruses. 2020; 12(10):1120. https://doi.org/10.3390/v12101120
Chicago/Turabian StyleHooper, Chantelle, Partho P. Debnath, Sukumar Biswas, Ronny van Aerle, Kelly S. Bateman, Siddhawartha K. Basak, Muhammad M. Rahman, Chadag V. Mohan, H. M. Rakibul Islam, Stuart Ross, and et al. 2020. "A Novel RNA Virus, Macrobrachium rosenbergii Golda Virus (MrGV), Linked to Mass Mortalities of the Larval Giant Freshwater Prawn in Bangladesh" Viruses 12, no. 10: 1120. https://doi.org/10.3390/v12101120