Calibration of Methods for SARS-CoV-2 Environmental Surveillance: A Case Study from Northwest Tuscany
Abstract
:1. Introduction
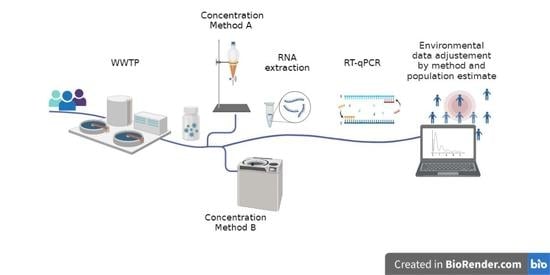
2. Materials and Methods
2.1. Environmental Surveillance for SARS-CoV-2
2.2. Samples Analysis for SARS-CoV-2 Genome
2.2.1. Sample Concentration Methods
2.2.2. SARS-CoV-2 RNA Extraction, Purification, and Detection
2.3. Lab-Scale Recovery Efficiency Assays
2.4. Clinical Data Source
2.5. Data Analysis
2.5.1. Virus Recovery Efficiency
2.5.2. Calibration between Analytical Methods
2.5.3. Normalization of SARS-CoV-2 Data
3. Results
3.1. SARS-CoV-2 Concentration in Sewage
3.2. Concentration Methods Efficiency
3.3. Results of the Calibration of Analytical Methods
3.4. SARS-CoV-2 Load after Adjustment by Method and Population
4. Discussion
5. Conclusions
- To assess the recovery efficiency of different analytical methods in order to adjust the data obtained with low sensitivity protocols;
- To normalize environmental viral concentrations by population in order to reduce potential errors attributable to wastewater volume or population size.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). Guidelines for Environmental Surveillance of Poliovirus Circulation Vaccines and Biologicals. Available online: https://polioeradication.org/wp-content/uploads/2016/07/WHO_V-B_03.03_eng.pdf (accessed on 12 October 2022).
- Carducci, A.; Morici, P.; Pizzi, F.; Battistini, R.; Rovini, E.; Verani, M. Study of the Viral Removal Efficiency in a Urban Wastewater Treatment Plant. Water Sci. Technol. 2008, 58, 893–897. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Feng, H.; Zhang, S.; Ni, Z.; Ni, L.; Chen, Y.; Zhuo, L.; Zhong, Z.; Qu, T. SARS-CoV-2 RNA Detection of Hospital Isolation Wards Hygiene Monitoring during the Coronavirus Disease 2019 Outbreak in a Chinese Hospital. Int. J. Infect. Dis. 2020, 94, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Jiang, X.; Zhang, Z.; Huang, S.; Zhang, Z.; Fang, Z.; Gu, Z.; Gao, L.; Shi, H.; Mai, L.; et al. Gastrointestinal Symptoms of 95 Cases with SARS-CoV-2 Infection. Gut 2020, 69, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Lescure, F.X.; Bouadma, L.; Nguyen, D.; Parisey, M.; Wicky, P.H.; Behillil, S.; Gaymard, A.; Bouscambert-Duchamp, M.; Donati, F.; le Hingrat, Q.; et al. Clinical and Virological Data of the First Cases of COVID-19 in Europe: A Case Series. Lancet Infect. Dis. 2020, 20, 697–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Chen, L.; Deng, Q.; Zhang, G.; Wu, K.; Ni, L.; Yang, Y.; Liu, B.; Wang, W.; Wei, C.; et al. The Presence of SARS-CoV-2 RNA in the Feces of COVID-19 Patients. J. Med. Virol. 2020, 92, 833–840. [Google Scholar] [CrossRef] [Green Version]
- Lo, I.L.; Lio, C.F.; Cheong, H.H.; Lei, C.I.; Cheong, T.H.; Zhong, X.; Tian, Y.; Sin, N.N. Evaluation of Sars-Cov-2 Rna Shedding in Clinical Specimens and Clinical Characteristics of 10 Patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020, 16, 1698–1707. [Google Scholar] [CrossRef] [Green Version]
- Foladori, P.; Cutrupi, F.; Segata, N.; Manara, S.; Pinto, F.; Malpei, F.; Bruni, L.; la Rosa, G. SARS-CoV-2 from Faeces to Wastewater Treatment: What Do We Know? A Review. Sci. Total Environ. 2020, 743, 140444. [Google Scholar] [CrossRef]
- World Health Organization (WHO) Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 12 October 2022).
- Centers for Disease Control and Prevention (CDC). National Wastewater Surveillance System (NWSS)-a New Public Health Tool to Understand COVID-19 Spread in a Community. Available online: https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/wastewater-surveillance.html#how-wastewater-surveillance-works (accessed on 12 October 2022).
- European Commission (EU). Commission Recommendation (EU) 2021/472 of 17 March 2021 on a Common Approach to Establish a Systematic Surveillance of SARS-CoV-2 and its Variants in Wastewaters in the EU-Publications Office of the EU. Available online: https://op.europa.eu/en/publication-detail/-/publication/05b46cb0-8855-11eb-ac4c-01aa75ed71a1/language-en/format-PDF (accessed on 12 October 2022).
- Li, X.; Zhang, S.; Shi, J.; Luby, S.P.; Jiang, G. Uncertainties in Estimating SARS-CoV-2 Prevalence by Wastewater-Based Epidemiology. Chem. Eng. J. 2021, 415, 129039. [Google Scholar] [CrossRef]
- Istituto Superiore di Sanità (ISS). Press Release N°41/2020-Wastewater and Coronavirus, the ‘Sentinel’ Network of Epidemiological Surveillance is Underway. CS N°41/2020-Acque Reflue e Coronavirus, Al Via La Rete ‘Sentinella’ Di Sorveglianza Epidemiologica Italy. Available online: https://www.iss.it/coronavirus/-/asset_publisher/1SRKHcCJJQ7E/content/id/5428743 (accessed on 2 December 2022). (In Italian).
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 937–942. [Google Scholar] [CrossRef]
- Pastorino, B.; Touret, F.; Gilles, M.; de Lamballerie, X.; Charrel, R.N. Heat inactivation of different types of SARS-CoV-2 samples: What protocols for biosafety, molecular detection and serological diagnostics? Viruses 2020, 12, 735. [Google Scholar] [CrossRef]
- La Rosa, G.; Iaconelli, M.; Bonadonna, L.; Suffredini, E. Protocollo Della Sorveglianza Di SARS-CoV-2 in Reflui Urbani (SARI)-Rev. 1. 2020. Available online: https://zenodo.org/record/5758709#.Y42R9PfP2Uk (accessed on 31 October 2022). [CrossRef]
- Wu, F.; Zhang, J.; Xiao, A.; Gu, X.; Lee, W.L.; Armas, F.; Kauffman, K.; Hanage, W.; Matus, M.; Ghaeli, N.; et al. SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases. mSystems 2020, 5, e00614-20. [Google Scholar] [CrossRef]
- La Rosa, G.; Iaconelli, M.; Bonadonna, L.; Suffredini, E. Protocollo Della Sorveglianza Di SARS-CoV-2 in Reflui Urbani (SARI)-Rev. 2. 2021. Available online: https://zenodo.org/record/5758714#.Y42et4czaUk (accessed on 31 October 2022). [CrossRef]
- La Rosa, G.; Mancini, P.; Bonanno Ferraro, G.; Veneri, C.; Iaconelli, M.; Bonadonna, L.; Lucentini, L.; Suffredini, E. SARS-CoV-2 Has Been Circulating in Northern Italy since December 2019: Evidence from Environmental Monitoring. Sci. Total Environ. 2021, 750, 141711. [Google Scholar] [CrossRef]
- ISO-ISO 15216-2:2019- Microbiology of the food chain—Horizontal method for Determination of Hepatitis A Virus and Norovirus Using Real-Time RT-PCR—Part 2: Method for Detection. Available online: https://www.iso.org/standard/74263.html (accessed on 12 October 2022).
- Yaniv, K.; Shagan, M.; Kramarsky-Winter, E.; Weil, M.; Indenbaum, V.; Elul, M.; Erster, O.; Brown, A.S.; Mendelson, E.; Mannasse, B.; et al. City-Level SARS-CoV-2 Sewage Surveillance. medRxiv 2021, arXiv:10.19.20215244. [Google Scholar] [CrossRef]
- Jevšnik, M.; Steyer, A.; Zrim, T.; Pokorn, M.; Mrvič, T.; Grosek, Š.; Strle, F.; Lusa, L.; Petrovec, M. Detection of Human Coronaviruses in Simultaneously Collected Stool Samples and Nasopharyngeal Swabs from Hospitalized Children with Acute Gastroenteritis. Virol. J. 2013, 10, 46. [Google Scholar] [CrossRef] [Green Version]
- European Center for Disease Prevention and Control (ECDC). COVID-19 Variant Dashboard (Beta). Available online: https://gis.ecdc.europa.eu/portal/apps/opsdashboard/index.html#/25b6e879c076412aaa9ae7adb78d3241 (accessed on 12 October 2022).
- Maida, C.M.; Amodio, E.; Mazzucco, W.; la Rosa, G.; Lucentini, L.; Suffredini, E.; Palermo, M.; Andolina, G.; Iaia, F.R.; Merlo, F.; et al. Wastewater-Based Epidemiology for Early Warning of SARS-COV-2 Circulation: A Pilot Study Conducted in Sicily, Italy. Int. J. Hyg. Environ. Health 2022, 242, 113948. [Google Scholar] [CrossRef]
- De Giglio, O.; Triggiano, F.; Apollonio, F.; Diella, G.; Fasano, F.; Stefanizzi, P.; Lopuzzo, M.; Brigida, S.; Calia, C.; Pousis, C.; et al. Potential Use of Untreated Wastewater for Assessing COVID-19 Trends in Southern Italy. Int. J. Environ. Res. Public Health 2021, 18, 10278. [Google Scholar] [CrossRef]
- Barril, P.A.; Pianciola, L.A.; Mazzeo, M.; Ousset, M.J.; Jaureguiberry, M.V.; Alessandrello, M.; Sánchez, G.; Oteiza, J.M. Evaluation of Viral Concentration Methods for SARS-CoV-2 Recovery from Wastewaters. Sci. Total Environ. 2021, 756, 144105. [Google Scholar] [CrossRef]
- Kumar, M.; Kuroda, K.; Patel, A.K.; Patel, N.; Bhattacharya, P.; Joshi, M.; Joshi, C.G. Decay of SARS-CoV-2 RNA along the Wastewater Treatment Outfitted with Upflow Anaerobic Sludge Blanket (UASB) System Evaluated through Two Sample Concentration Techniques. Sci. Total Environ. 2021, 754, 142329. [Google Scholar] [CrossRef]
- Mahon, J.M.; Javier, A.; Monleon, C.; Gill, L.W.; O’sullivan, J.J.; Meijer, W.G. Wastewater-Based Epidemiology (WBE) for SARS-CoV-2-A Review Focussing on the Significance of the Sewer Network Using a Dublin City Catchment Case Study. Water Sci. Technol. 2022, 86, 1402–1425. [Google Scholar] [CrossRef]
- Wade, M.J.; lo Jacomo, A.; Armenise, E.; Brown, M.R.; Bunce, J.T.; Cameron, G.J.; Fang, Z.; Farkas, K.; Gilpin, D.F.; Graham, D.W.; et al. Understanding and Managing Uncertainty and Variability for Wastewater Monitoring beyond the Pandemic: Lessons Learned from the United Kingdom National COVID-19 Surveillance Programmes. J. Hazard. Mater. 2022, 424, 127456. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, M.; Iker, B.C.; Pepper, I.L.; Gerba, C.P. Relative Abundance and Treatment Reduction of Viruses during Wastewater Treatment Processes—Identification of Potential Viral Indicators. Sci. Total Environ. 2014, 488–489, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Maal-Bared, R.; Qiu, Y.; Li, Q.; Gao, T.; Hrudey, S.E.; Bhavanam, S.; Ruecker, N.J.; Ellehoj, E.; Lee, B.E.; Pang, X. Does Normalization of SARS-CoV-2 Concentrations by Pepper Mild Mottle Virus Improve Correlations and Lead Time between Wastewater Surveillance and Clinical Data in Alberta (Canada): Comparing Twelve SARS-CoV-2 Normalization Approaches. Sci. Total Environ. 2023, 856, 158964. [Google Scholar] [CrossRef] [PubMed]





| Virus (Target Region) | Primers and Probes Name | Concentrations (μM) | Sequences (5′-3′) | Thermal Amplification Conditions |
|---|---|---|---|---|
| CoV-2-F | 0.5 | ACA TGG CTT TGA GTT GAC ATC T | ||
| SARS-CoV-2 (ORF-1ab region) | CoV-2-R | 0.9 | AGC AGT GGA AAA GCAT GTG G | 50°C: 30 min, 95 °C: 5 min, 45 cycles (95 °C: 15 s; 60°C: 45 s) |
| CoV-2-P | 0.25 | FAM—CAT AGA CAA CAG GTG CGC TC-MGBEQ | ||
| HCV 229E-F | 0.5 | GAT GCA ACT ACA GCC TAC GC | ||
| Human Coronavirus 229E (ORF-1ab) | HCV 229E-R | 0.9 | AGT TAA CGC TCA AAA CGC AAT | 50°C: 30 min, 95 °C: 5 min, 45 cycles (95 °C: 15 s; 60°C: 30 s) |
| HCV 229E—P | 0.25 | FAM—TTT CAG GCT GTA AGT TCT AAC ATT-TAMRA | ||
| Mengo -F | 0.5 | GCG GGT CCT GCC GAA AGT | ||
| Mengovirus (5′UTR [21]) | Mengo -R | 0.9 | GAA GTA ACA TAT AGA CAG ACG CAC AC | 45 °C: 10 min, 95 °C: 10 min, 45 cycles (95 °C: 15 s; 60 °C: 45 s) |
| Mengo -P | 0.2 | FAM—ATC ACA TTA CTG GCC GAA GC- MGBNFQ |
| Replicates | Method A | Method B | ||
|---|---|---|---|---|
| Concentration (GC/µL) | Recovery Rate (%) | Concentration (GC/µL) | Recovery Rate (%) | |
| vMC0 (spike = 2.11 × 106 GC/µL) | ||||
| 1 | 2.74 × 103 | 0.13 | 2.46 × 105 | 11.65 |
| 2 | 1.36 × 104 | 0.64 | 5.54 × 105 | 26.25 |
| 3 | 5.34 × 103 | 0.25 | 6.21 × 105 | 29.43 |
| 4 | 9.73 × 103 | 0.92 | 3.47 × 105 | 16.44 |
| HCoV-229E (spike = 6.31 × 1010 GC/µL) | ||||
| 1 | 1.18 × 109 | 1.87 | 1.78 × 1010 | 28.20 |
| 2 | 2.10 × 109 | 3.32 | 1.60 × 1010 | 25.35 |
| 3 | 1.53 × 109 | 2.42 | 6.43 × 109 | 10.19 |
| 4 | 1.64 × 109 | 2.60 | 1.09 × 109 | 1.73 |
| Name of WWTPs | Inflow Rate (Median Value) | Population Equivalent |
|---|---|---|
| WWTP1 | 10,128 | 42,931 |
| WWTP2 | 13,368 | 68,070 |
| WWTP3 | 20,071 | 110,871 |
| WWTP4 | 11,378 | 60,262 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verani, M.; Federigi, I.; Muzio, S.; Lauretani, G.; Calà, P.; Mancuso, F.; Salvadori, R.; Valentini, C.; La Rosa, G.; Suffredini, E.; et al. Calibration of Methods for SARS-CoV-2 Environmental Surveillance: A Case Study from Northwest Tuscany. Int. J. Environ. Res. Public Health 2022, 19, 16588. https://doi.org/10.3390/ijerph192416588
Verani M, Federigi I, Muzio S, Lauretani G, Calà P, Mancuso F, Salvadori R, Valentini C, La Rosa G, Suffredini E, et al. Calibration of Methods for SARS-CoV-2 Environmental Surveillance: A Case Study from Northwest Tuscany. International Journal of Environmental Research and Public Health. 2022; 19(24):16588. https://doi.org/10.3390/ijerph192416588
Chicago/Turabian StyleVerani, Marco, Ileana Federigi, Sara Muzio, Giulia Lauretani, Piergiuseppe Calà, Fabrizio Mancuso, Roberto Salvadori, Claudia Valentini, Giuseppina La Rosa, Elisabetta Suffredini, and et al. 2022. "Calibration of Methods for SARS-CoV-2 Environmental Surveillance: A Case Study from Northwest Tuscany" International Journal of Environmental Research and Public Health 19, no. 24: 16588. https://doi.org/10.3390/ijerph192416588