Photoinduced Synthesis of Methylated Marine Cyclopeptide Galaxamide Analogs with Isoindolinone as Anticancer Agents
Abstract
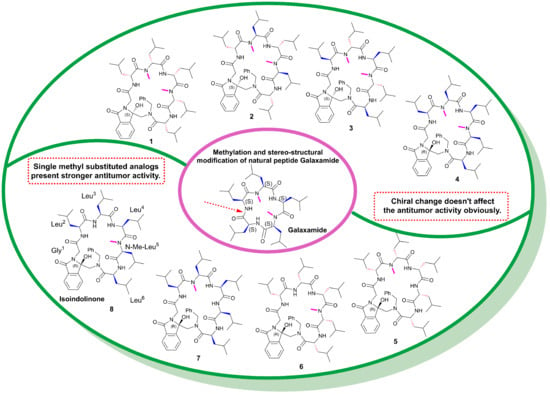
:1. Introduction
2. Results and Discussion
2.1. Synthesis
2.2. Determination of Absolute Configurations (ACs)
2.3. MTT Method to Detect the Effect of Drugs on Cell Proliferation
2.4. Apoptosis Detection by Flow Cytometry Analysis
2.5. Effect of Compound 8 on Cell Cycle
2.6. Effect of Compound 8 on Mitochondrial Membrane Potential
2.7. Effect of Compound 8 on Intracellular Ca2+ Levels and Lactate Dehydrogenase Activity
2.8. The Influence on MDM2 and p53 Expression of Compound 8
2.9. Molecular Docking to MDM2 Protein
3. Materials and Methods
3.1. General
3.2. Synthesis of the Photoreaction Precursor Linear Peptides (9–16)
3.2.1. N-Phthalimido-Gly-Leu-N-Me-Leu-Leu-N-Me-Leu-Leu-Si(CH3)3 (9)
3.2.2. N-Phthalimido-Gly-Leu-N-Me-Leu-Leu-N-Me-Leu-Leu-Si(CH3)3 (10)
3.2.3. N-Phthalimido-Gly-Leu-N-Me-Leu-Leu-N-Me-Leu-Leu-Si(CH3)3 (11)
3.2.4. N-Phthalimido-Gly-Leu-N-Me-Leu-Leu-N-Me-Leu-Leu-Si(CH3)3 (12)
3.2.5. N-Phthalimido-Gly-Leu-Leu-Leu-N-Me-Leu-Leu-Si(CH3)3 (13)
3.2.6. N-Phthalimido-Gly-Leu-N-Me-Leu-Leu-Leu-Leu-Si(CH3)3 (14)
3.2.7. N-Phthalimido-Gly-Leu-Leu-Leu-N-Me-Leu-Leu-Si(CH3)3 (15)
3.2.8. N-Phthalimido-Gly-Leu-N-Me-Leu-Leu-Leu-Leu-Si(CH3)3 (16)
3.3. Preparation of Cyclic Peptides 1–8
3.3.1. 3-Hydroxy-isoindolinone-cyclo-(Gly-Leu-N-Me-Leu-Leu-N-Me-Leu-Leu) (1)
3.3.2. 3-Hydroxy-isoindolinone-cyclo-Gly-Leu-N-Me-Leu-Leu-N-Me-Leu-Leu (2)
3.3.3. 3-Hydroxy-isoindolinone-cyclo-Gly-Leu-N-Me-Leu-Leu-N-Me-Leu-Leu (3)
3.3.4. 3-Hydroxy-isoindolinone-cyclo-Gly-Leu-N-Me-Leu-Leu-N-Me-Leu-Leu (4)
3.3.5. 3-Hydroxy-isoindolinone-cyclo-Gly-Leu-Leu-Leu-N-Me-Leu-Leu (5)
3.3.6. 3-Hydroxy-isoindolinone-cyclo-Gly-Leu-N-Me-Leu-Leu-Leu-Leu (6)
3.3.7. 3-Hydroxy-isoindolinone-cyclo-Gly-Leu-Leu-Leu-N-Me-Leu-Leu (7)
3.3.8. 3-Hydroxy-isoindolinone-cyclo-Gly-Leu-N-Me-Leu-Leu-Leu-Leu (8)
3.4. MTT Experiment
3.5. Detecting Apoptosis by Flow Cytometry
3.6. Detecting the Effect on the Cell Cycle by Flow Cytometry
3.7. Detecting the Effect on Mitochondrial Membrane Potential by Flow Cytometry
3.8. Detecting the Effect on Intracellular Ca2+ Concentration
3.9. Lactate Dehydrogenase (LDH) Activity Assay
3.10. Effect on Protein Levels of MDM2 and p53 by Western Blot
3.11. Theoretical Computational Details
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daly, N.L.; Wilson, D.T. Plant derived cyclic peptides. Biochem. Soc. Trans. 2021, 49, 1279–1285. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, S. Cyclic peptide drugs approved in the last two decades (2001–2021). RSC Chem. Biol. 2021, 3, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, A.A.; Yin, Y.; Suga, H. Macrocyclic Peptides as Drug Candidates: Recent Progress and Remaining Challenges. J. Am. Chem. Soc. 2019, 141, 4167–4181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-N.; Xia, Y.-X.; Zhang, H.-J. Natural Cyclopeptides as Anticancer Agents in the Last 20 Years. Int. J. Mol. Sci. 2021, 22, 3973. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.-J.; Liao, X.-J.; Xu, S.-H.; Diao, J.-Z.; Du, B.; Zhou, X.-L.; Pan, S.-S. ChemInform Abstract: Isolation, Structure Determination, and Synthesis of Galaxamide, a Rare Cytotoxic Cyclic Pentapeptide from a Marine Algae Galaxaura filamentosa. Org. Lett. 2008, 10, 4569–4572. [Google Scholar] [CrossRef]
- Xiao, X.; Liao, X.; Qiu, S.; Liu, Z.; Du, B.; Xu, S. Synthesis, cytotoxicity and apoptosis induction in human tumor cells by galaxamide and its analogues. Mar Drugs. 2014, 12, 4521–4538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunagariya, J.; Zhong, S.; Chen, J.; Bai, D.; Bhadja, P.; Long, W.; Liao, X.; Tang, X.; Xu, S. Design and synthesis of analogues of marine natural product gtalaxamide, an N-methylated cyclic pentapeptide, as potential anti-yumor agent in vitro. Mar. Drugs 2016, 14, 161. [Google Scholar] [CrossRef] [Green Version]
- Lunagariya, J.; Liao, X.; Long, W.; Zhong, S.; Bhadja, P.; Li, H.; Zhao, B.; Xu, S. Cytotoxicity Study of Cyclopentapeptide Analogues of Marine Natural Product Galaxamide towards Human Breast Cancer Cells. Oxidative Med. Cell. Longev. 2017, 2017, 8392035. [Google Scholar] [CrossRef]
- Bai, D.; Yu, S.; Zhong, S.; Zhao, B.; Qiu, S.; Chen, J.; Lunagariya, J.; Liao, X.; Xu, S. d-Amino Acid Position Influences the Anticancer Activity of Galaxamide Analogs: An Apoptotic Mechanism Study. Int. J. Mol. Sci. 2017, 18, 544. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Gu, W.; Lo, D.; Ding, X.-Z.; Ujiki, M.; Adrian, T.E.; Soff, G.A.; Silverman, R.B. N-Methylsansalvamide A Peptide Analogues. Potent New Antitumor Agents. J. Med. Chem. 2005, 48, 3630–3638. [Google Scholar] [CrossRef]
- Li, Y.; Li, W.; Xu, Z. Improvement on Permeability of Cyclic Peptide/Peptidomimetic: Backbone N-Methylation as A Useful Tool. Mar. Drugs 2021, 19, 311. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhao, L.; Wang, Z.; Jin, Y. Photo-induced synthesis of chiral galaxamide analogs and the biological activities against human tumor cells. New J. Chem. 2018, 42, 19779–19784. [Google Scholar] [CrossRef]
- Zhao, L.; Wu, J.; Bao, Y.; Jiang, S.; Wang, Z.; Guo, C.; Jin, Y.; Qu, F. Synthesis of novel isoindole-containing phakellistatin 2 analogs and the conformation features affecting their antitumor activities. New J. Chem. 2019, 43, 12609–12613. [Google Scholar] [CrossRef]
- Schilling, N.A.; Berscheid, A.; Schumacher, J.; Saur, J.S.; Konnerth, M.C.; Wirtz, S.N.; Beleña, J.M.B.; Zipperer, A.; Krismer, B.; Peschel, A.; et al. Synthetic Lugdunin Analogues Reveal Essential Structural Motifs for Antimicrobial Action and Proton Translocation Capability. Angew. Chem. Int. Ed. 2019, 58, 9234–9238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grauso, L.; Teta, R.; Esposito, G.; Menna, M.; Mangoni, A. Computational prediction of chiroptical properties in structure elu-cidation of natural products. Nat. Prod. Rep. 2019, 36, 1005–1030. [Google Scholar] [CrossRef]
- Migliore, M.; Bonvicini, A.; Tognetti, V.; Guilhaudis, L.; Baaden, M.; Oulyadi, H.; Joubert, L.; Ségalas-Milazzo, I. Characterization of β-turns by electronic circular dichroism spectroscopy: A coupled molecular dynamics and time-dependent density functional theory computational study. Phys. Chem. Chem. Phys. 2020, 22, 1611–1623. [Google Scholar] [CrossRef]
- Kumar, A.; Schweitzer-Stenner, R.; Wong, B.M. A new interpretation of the structure and solvent dependence of the far UV circular dichroism spectrum of short oligopeptides. Chem. Commun. 2019, 55, 5701–5704. [Google Scholar] [CrossRef]
- Kronik, L.; Stein, T.; Refaely-Abramson, S.; Baer, R. Water-mediated electronic structure of oligopeptides probed by their UV circular dichroism, absorption spectra, and time-dependent DFT calculations. J. Chem. Theory Comput. 2012, 8, 1515–1531. [Google Scholar] [CrossRef]
- Kaufmann, S.H.; Earnshaw, W. Induction of Apoptosis by Cancer Chemotherapy. Exp. Cell Res. 2000, 256, 42–49. [Google Scholar] [CrossRef]
- Guo, M.; Lu, B.; Gan, J.; Wang, S.; Jiang, X.; Li, H. Apoptosis detection: A purpose-dependent approach selection. Cell Cycle 2021, 20, 1033–1040. [Google Scholar] [CrossRef]
- Ormerod, M.G. Investigating the relationship between the cell cycle and apoptosis using flow cytometry. J. Immunol. Methods 2002, 265, 73–80. [Google Scholar] [CrossRef]
- Kuwahara, Y.; Tomita, K.; Roudkenar, M.H.; Roushandeh, A.M.; Urushihara, Y.; Igarashi, K.; Kurimasa, A.; Sato, T. Decreased mitochondrial membrane potential is an indicator of radioresistant cancer cells. Life Sci. 2021, 286, 120051. [Google Scholar] [CrossRef] [PubMed]
- Klier, P.E.Z.; Martin, J.G.; Miller, E.W. Imaging Reversible Mitochondrial Membrane Potential Dynamics with a Masked Rhodamine Voltage Reporter. J. Am. Chem. Soc. 2021, 143, 4095–4099. [Google Scholar] [CrossRef] [PubMed]
- Orrenius, S.; Zhivotovsky, B.; Nicotera, P. Regulation of cell death: The calcium–apoptosis link. Nat. Rev. Mol. Cell Biol. 2003, 4, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Oldach, L. Democratizing calcium visualization. J. Biol. Chem. 2021, 297, 101181. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell pro-liferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [Green Version]
- Szabo, I.; Zoratti, M.; Biasutto, L. Targeting mitochondrial ion channels for cancer therapy. Redox Biol. 2021, 42, 101846. [Google Scholar] [CrossRef]
- Duffy, M.J.; Synnott, N.C.; McGowan, P.M.; Crown, J.; O’Connor, D.; Gallagher, W. p53 as a target for the treatment of cancer. Cancer Treat. Rev. 2014, 40, 1153–1160. [Google Scholar] [CrossRef]
- Li, Y.; Yang, J.; Aguilar, A.; McEachern, D.; Przybranowski, S.; Liu, L.; Yang, C.-Y.; Wang, M.; Han, X.; Wang, S. Discovery of MD-224 as a First-in-Class, Highly Potent, and Efficacious Proteolysis Targeting Chimera Murine Double Minute 2 Degrader Capable of Achieving Complete and Durable Tumor Regression. J. Med. Chem. 2019, 62, 448–466. [Google Scholar] [CrossRef]
- Mullard, A. p53 programmes plough on. Nat. Rev. Drug Discov. 2020, 19, 497–500. [Google Scholar] [CrossRef]
- Baek, S.; Kutchukian, P.S.; Verdine, G.L.; Huber, R.; Holak, T.A.; Lee, K.W.; Popowicz, G.M. Structure of the Stapled p53 Peptide Bound to Mdm2. J. Am. Chem. Soc. 2012, 134, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.S.; Graves, B.; Guerlavais, V.; Tovar, C.; Packman, K.; To, K.H.; Olson, K.A.; Kesavan, K.; Gangurde, P.; Mukherjee, A.; et al. Stapled α−helical peptide drug development: A potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. USA 2013, 110, E3445–E3454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Tolbert, W.D.; Hu, H.-G.; Gohain, N.; Zou, Y.; Niu, F.; He, W.-X.; Yuan, W.; Su, J.-C.; Pazgier, M.; et al. Dithiocarbamate-inspired side chain stapling chemistry for peptide drug design. Chem. Sci. 2019, 10, 1522–1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giofrè, S.V.; Cirmi, S.; Mancuso, R.; Nicolò, F.; Lanza, G.; Legnani, L.; Campisi, A.; Chiacchio, M.A.; Navarra, M.; Gabriele, B. Synthesis of spiro[isoindole-1,5’-isoxazolidin]-3(2H)-ones as potential inhibitors of the MDM2-p53 interaction. Beilstein J. Org. Chem. 2016, 12, 2793–2807. [Google Scholar] [CrossRef] [Green Version]
- Hardcastle, I.R.; Liu, J.; Valeur, E.; Watson, A.; Ahmed, S.U.; Blackburn, T.J.; Bennaceur, K.; Clegg, W.; Drummond, C.; Endicott, J.A.; et al. Isoindolinone Inhibitors of the Murine Double Minute 2 (MDM2)-p53 Protein−Protein Interaction: Structure−Activity Studies Leading to Improved Potency. J. Med. Chem. 2011, 54, 1233–1243. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, X.; Li, G.; Liu, X. Molecular Mechanism of Huaihuasan in treatment of Ulcerative Colitis based on network pharmacology and molecular docking. Phytomed. Plus 2021, 1, 100081. [Google Scholar] [CrossRef]
- Bao, Y.; Zhao, L.; Wu, J.; Jiang, S.; Wang, Z.; Jin, Y. Photo-induced synthesis of Axinastatin 3 analogs, the secondary structures and their in vitro antitumor activities. Bioorg. Med. Chem. Lett. 2019, 29, 126730. [Google Scholar] [CrossRef]
- Bruhn, T.; Schaumlöffel, A.; Hemberger, Y.; Pescitelli, G. SpecDis, Version 1.71; University of Wuerzburg: Wuerzburg, Germany, 2017.
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]






| Compounds | IC50 (μM) | ||
|---|---|---|---|
| HepG-2 | MDA-MB-231 | L929 [a]p | |
| 1 | 38.4 ± 2.2 | 59.3 ± 2.3 | >400 |
| 2 | 72.1 ± 5.8 | 67.6 ± 3.3 | 363.6 ± 15.6 |
| 3 | 90.9 ± 7.69 | 73.2 ± 2.8 | >400 |
| 4 | 80.99 ± 4.29 | 95.3 ± 8.6 | 310.4 ± 20.1 |
| 5 | 28.2 ± 6.2 | 60.8 ± 8.2 | 153.1 ± 10.7 |
| 6 | 36.1 ± 2.2 | 67.6 ± 2.3 | 120.5 ± 5.3 |
| 7 | 30.4 ± 1.6 | 111.5 ± 9.0 | >400 |
| 8 | 11.1± 0.7 | 41.7 ± 3.2 | >400 |
| Taxol | 40.9 ± 5.2 | - | 140.6 ± 9.3 |
| Compounds | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| EBinding | −5.59 | −5.87 | −5.91 | −5.5 | −5.2 | −6.43 | −6.7 | −10.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, S.; Wang, Z.; Zhang, H.; Zhao, L.; Chang, Q.; Zhang, X.; Yan, R.; Wu, X.; Jin, Y. Photoinduced Synthesis of Methylated Marine Cyclopeptide Galaxamide Analogs with Isoindolinone as Anticancer Agents. Mar. Drugs 2022, 20, 379. https://doi.org/10.3390/md20060379
Xiao S, Wang Z, Zhang H, Zhao L, Chang Q, Zhang X, Yan R, Wu X, Jin Y. Photoinduced Synthesis of Methylated Marine Cyclopeptide Galaxamide Analogs with Isoindolinone as Anticancer Agents. Marine Drugs. 2022; 20(6):379. https://doi.org/10.3390/md20060379
Chicago/Turabian StyleXiao, Shimei, Zhiqiang Wang, Huanli Zhang, Lei Zhao, Qingran Chang, Xiong Zhang, Rui Yan, Xiaodan Wu, and Yingxue Jin. 2022. "Photoinduced Synthesis of Methylated Marine Cyclopeptide Galaxamide Analogs with Isoindolinone as Anticancer Agents" Marine Drugs 20, no. 6: 379. https://doi.org/10.3390/md20060379