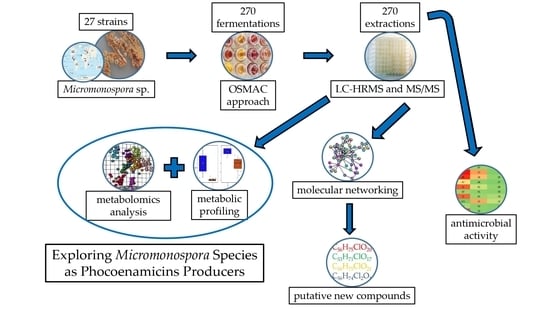
Exploring Micromonospora as Phocoenamicins Producers
Abstract
:1. Introduction
2. Results
2.1. Taxonomy, Geography and Ecology of the Producing Microorganisms
2.2. OSMAC Approach, Extraction and Metabolomics Analysis of the Fermentation Broths
2.3. Antimicrobial Activity of the Crude Extracts
2.4. Dereplication of The Main Compounds Produced
2.5. Molecular Networking Analysis
2.6. Putative Identification of Possible New Compounds
3. Discussion
4. Materials and Methods
4.1. Taxonomical Identification of the Producing Microorganisms
4.2. Fermentation of the Producing Microorganisms and OSMAC Approach
4.3. Extraction of the Fermentation Broths and Analysis by LC-HRMS
4.4. Multivariate Data Analysis (MVDA) with MZmine 2 and MetaboAnalyst 5.0
4.5. High-Throughput Screening Assay of the Fermentation Extracts for Antimicrobial Activities
4.6. Molecular Networking
4.6.1. Classical Molecular Networking
4.6.2. Preprocessing by MZmine 2 and Feature-Based Molecular Networking
4.7. Dereplication Process and Identification of Putative New Analogues
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adebisi, Y.A.; Alaran, A.J.; Okereke, M.; Oke, G.I.; Amos, O.A.; Olaoye, O.C.; Oladunjoye, I.; Olanrewaju, A.Y.; Ukor, N.A.; Lucero-Prisno, D.E. COVID-19 and Antimicrobial Resistance: A Review. Infect. Dis. Res. Treat. 2021, 14, 117863372110338. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Davison, E.K.; Brimble, M.A. Natural product derived privileged scaffolds in drug discovery. Curr. Opin. Chem. Biol. 2019, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O. Natural products discovery and potential for new antibiotics. Curr. Opin. Microbiol. 2019, 51, 81–87. [Google Scholar] [CrossRef]
- Niu, G.; Li, W. Next-Generation Drug Discovery to Combat Antimicrobial Resistance. Trends Biochem. Sci. 2019, 44, 961–972. [Google Scholar] [CrossRef]
- De Mol, M.L.; Snoeck, N.; De Maeseneire, S.L.; Soetaert, W.K. Hidden antibiotics: Where to uncover? Biotechnol. Adv. 2018, 36, 2201–2218. [Google Scholar] [CrossRef]
- Mohana, C.; Rao, H.Y.; Rakshith, D.; Mithun, P.; Nuthan, B.R.; Satish, S. Omics based approach for biodiscovery of microbial natural products in antibiotic resistance era. J. Genet. Eng. Biotechnol. 2018, 16, 1–8. [Google Scholar] [CrossRef]
- Abdel-Razek, A.S.; El-Naggar, M.E.; Allam, A.; Morsy, O.M.; Othman, S.I. Microbial Natural Products in Drug Discovery. Processes 2020, 8, 470. [Google Scholar] [CrossRef]
- Quinn, R.A.; Nothias, L.-F.; Vining, O.; Meehan, M.; Esquenazi, E.; Dorrestein, P.C. Molecular Networking As a Drug Discovery, Drug Metabolism, and Precision Medicine Strategy. Trends Pharmacol. Sci. 2017, 38, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Lacoske, M.H.; Theodorakis, E.A. Spirotetronate Polyketides as Leads in Drug Discovery. J. Nat. Prod. 2015, 78, 562–575. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.; Maffioli, S.I.; Gaspari, E.; Rossi, R.; Mauri, P.; Sosio, M.; Donadio, S. Chrolactomycins from the Actinomycete Actinospica. J. Nat. Prod. 2012, 75, 1991–1993. [Google Scholar] [CrossRef]
- Braddock, A.A.; Theodorakis, E.A. Marine Spirotetronates: Biosynthetic Edifices That Inspire Drug Discovery. Mar. Drugs 2019, 17, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Bonilla, M.; Oves-Costales, D.; de la Cruz, M.; Kokkini, M.; Martín, J.; Vicente, F.; Genilloud, O.; Reyes, F. Phocoenamicins B and C, New Antibacterial Spirotetronates Isolated from a Marine Micromonospora sp. Mar. Drugs 2018, 16, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochoa, J.L.; Sanchez, L.M.; Koo, B.-M.; Doherty, J.S.; Rajendram, M.; Huang, K.C.; Gross, C.A.; Linington, R.G. Marine Mammal Microbiota Yields Novel Antibiotic with Potent Activity Against Clostridium difficile. ACS Infect. Dis. 2018, 4, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Victoria, I.; Martín, J.; Reyes, F. Combined LC/UV/MS and NMR Strategies for the Dereplication of Marine Natural Products. Planta Med. 2016, 82, 857–871. [Google Scholar] [CrossRef] [Green Version]
- Demain, A.L.; Gómez-Ortiz, B.; Ruiz-Villafán, B.; Rodríguez-Sanoja, R.; Sánchez, S. Recent findings of molecules with anti-infective activity: Screening of non-conventional sources. Curr. Opin. Pharmacol. 2019, 48, 40–47. [Google Scholar] [CrossRef]
- Ossai, J.; Khatabi, B.; Nybo, S.E.; Kharel, M.K. Renewed interests in the discovery of bioactive actinomycete metabolites driven by emerging technologies. J. Appl. Microbiol. 2022, 132, 59–77. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big Effects from Small Changes: Possible Ways to Explore Nature’s Chemical Diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Lever, J.; Krzywinski, M.; Altman, N. Points of Significance: Principal component analysis. Nat. Methods 2017, 14, 641–642. [Google Scholar] [CrossRef] [Green Version]
- Worley, B. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2012, 1, 92–107. [Google Scholar] [CrossRef]
- Ruiz-Perez, D.; Guan, H.; Madhivanan, P.; Mathee, K.; Narasimhan, G. So you think you can PLS-DA? BMC Bioinform. 2020, 21, 2. [Google Scholar] [CrossRef]
- Chen, Y.; Li, E.-M.; Xu, L.-Y. Guide to Metabolomics Analysis: A Bioinformatics Workflow. Metabolites 2022, 12, 357. [Google Scholar] [CrossRef]
- Caesar, L.K.; Kvalheim, O.M.; Cech, N.B. Hierarchical cluster analysis of technical replicates to identify interferents in untargeted mass spectrometry metabolomics. Anal. Chim. Acta 2018, 1021, 69–77. [Google Scholar] [CrossRef]
- Shamikh, Y.I.; El Shamy, A.A.; Gaber, Y.; Abdelmohsen, U.R.; Madkour, H.A.; Horn, H.; Hassan, H.M.; Elmaidomy, A.H.; Alkhalifah, D.H.M.; Hozzein, W.N. Actinomycetes from the Red Sea Sponge Coscinoderma mathewsi: Isolation, Diversity, and Potential for Bioactive Compounds Discovery. Microorganisms 2020, 8, 783. [Google Scholar] [CrossRef]
- Thamchaipenet, A. Maklamicin, an Antibacterial Polyketide from an Endophytic Micromonospora sp. J. Nat. Prod. 2011, 74, 670–674. [Google Scholar]
- Holden, P.; Nair, L.S. Deferoxamine: An Angiogenic and Antioxidant Molecule for Tissue Regeneration. Tissue Eng. Part B Rev. 2019, 25, 461–470. [Google Scholar] [CrossRef] [Green Version]
- Normant, V.; Josts, I.; Kuhn, L.; Perraud, Q.; Fritsch, S.; Hammann, P.; Mislin, G.L.A.; Tidow, H.; Schalk, I.J. Nocardamine-Dependent Iron Uptake in Pseudomonas aeruginosa: Exclusive Involvement of the FoxA Outer Membrane Transporter. ACS Chem. Biol. 2020, 15, 2741–2751. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-Y.; Graziani, E.; Waters, B.; Pan, W.; Li, X.; McDermott, J.; Meurer, G.; Saxena, G.; Andersen, R.J.; Davies, J. Novel Natural Products from Soil DNA Libraries in a Streptomycete Host. Org. Lett. 2000, 2, 2401–2404. [Google Scholar] [CrossRef] [PubMed]
- Iijima, M.; Someno, T.; Ishizuka, M.; Sawa, R.; Naganawa, H.; Takeuchi, T. IC202B and C, new siderophores with immunosuppressive activity produced by Streptoalloteichus sp. 1454-19. J. Antibiot. 1999, 52, 775–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Shaer, D.; Al Musaimi, O.; de la Torre, B.G.; Albericio, F. Hydroxamate siderophores: Natural occurrence, chemical synthesis, iron binding affinity and use as Trojan horses against pathogens. Eur. J. Med. Chem. 2020, 208, 112791. [Google Scholar] [CrossRef] [PubMed]
- Acquah, K.S.; Beukes, D.R.; Warner, D.F.; Meyers, P.R.; Sunassee, S.N.; Maglangit, F.; Deng, H.; Jaspars, M.; Gammon, D.W. Novel South African Rare Actinomycete Kribbella speibonae Strain SK5: A Prolific Producer of Hydroxamate Siderophores Including New Dehydroxylated Congeners. Molecules 2020, 25, 2979. [Google Scholar] [CrossRef] [PubMed]
- Bellotti, D.; Remelli, M. Deferoxamine B: A Natural, Excellent and Versatile Metal Chelator. Molecules 2021, 26, 3255. [Google Scholar] [CrossRef]
- Sun, M.-Y.; Ye, Y.; Xiao, L.; Rahman, K.; Xia, W.; Zhang, H. Daidzein: A review of pharmacological effects. Afr. J. Tradit. Complement. Altern. Med. 2016, 13, 117–132. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Quispe, C.; Imran, M.; Rauf, A.; Nadeem, M.; Gondal, T.A.; Ahmad, B.; Atif, M.; Mubarak, M.S.; Sytar, O.; et al. Genistein: An Integrative Overview of Its Mode of Action, Pharmacological Properties, and Health Benefits. Oxidative Med. Cell. Longev. 2021, 2021, 3268136. [Google Scholar] [CrossRef]
- Wang, J.-F.; Liu, S.-S.; Song, Z.-Q.; Xu, T.-C.; Liu, C.-S.; Hou, Y.-G.; Huang, R.; Wu, S.-H. Naturally Occurring Flavonoids and Isoflavonoids and Their Microbial Transformation: A Review. Molecules 2020, 25, 5112. [Google Scholar] [CrossRef]
- Ondrejíčková, P.; Šturdíková, M.; Hushegyi, A.; Švajdlenka, E.; Markošová, K.; Čertík, M. Endophytic Streptomyces sp. AC35, a producer of bioactive isoflavone aglycones and antimycins. J. Ind. Microbiol. Biotechnol. 2016, 43, 1333–1344. [Google Scholar] [CrossRef]
- Hamamoto, T.; Seto, H.; Beppu, T. Leptomycins A and B, new antifungal antibiotics. II. Structure elucidation. J. Antibiot. 1983, 36, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Afoullouss, S.; Balsam, A.; Allcock, A.L.; Thomas, O.P. Optimization of LC-MS2 Data Acquisition Parameters for Molecular Networking Applied to Marine Natural Products. Metabolites 2022, 12, 245. [Google Scholar] [CrossRef] [PubMed]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.-F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.-Y.; Tian, Z.-H.; Shao, L.; Qu, X.-D.; Zhao, Q.-F.; Tang, J.; Tang, G.-L.; Liu, W. Genetic Characterization of the Chlorothricin Gene Cluster as a Model for Spirotetronate Antibiotic Biosynthesis. Chem. Biol. 2006, 13, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Daduang, R.; Kitani, S.; Sudoh, Y.; Pait, I.G.U.; Thamchaipenet, A.; Ikeda, H.; Igarashi, Y.; Nihira, T. 29-Deoxymaklamicin, a new maklamicin analogue produced by a genetically engineered strain of Micromonospora sp. NBRC 110955. J. Biosci. Bioeng. 2015, 120, 608–613. [Google Scholar] [CrossRef]
- Wang, W.; Qiu, Z.; Tan, H.; Cao, L. Siderophore production by actinobacteria. BioMetals 2014, 27, 623–631. [Google Scholar] [CrossRef]
- Lacret, R.; Pérez-Victoria, I.; Oves-Costales, D.; de la Cruz, M.; Domingo, E.; Martín, J.; Díaz, C.; Vicente, F.; Genilloud, O.; Reyes, F. MDN-0170, a New Napyradiomycin from Streptomyces sp. Strain CA-271078. Mar. Drugs 2016, 14, 188. [Google Scholar] [CrossRef] [Green Version]
- Martín, J.; Crespo, G.; González-Menéndez, V.; Pérez-Moreno, G.; Sánchez-Carrasco, P.; Pérez-Victoria, I.; Ruiz-Pérez, L.M.; González-Pacanowska, D.; Vicente, F.; Genilloud, O.; et al. MDN-0104, an Antiplasmodial Betaine Lipid from Heterospora chenopodii. J. Nat. Prod. 2014, 77, 2118–2123. [Google Scholar] [CrossRef]
- Myers, O.D.; Sumner, S.J.; Li, S.; Barnes, S.; Du, X. One Step Forward for Reducing False Positive and False Negative Compound Identifications from Mass Spectrometry Metabolomics Data: New Algorithms for Constructing Extracted Ion Chromatograms and Detecting Chromatographic Peaks. Anal. Chem. 2017, 89, 8696–8703. [Google Scholar] [CrossRef]
- Audoin, C.; Bonhomme, D.; Ivanisevic, J.; De la Cruz, M.; Cautain, B.; Monteiro, M.C.; Reyes, F.; Rios, L.; Perez, T.; Thomas, O.P. Balibalosides, an Original Family of Glucosylated Sesterterpenes Produced by the Mediterranean Sponge Oscarella balibaloi. Mar. Drugs 2013, 11, 1477–1489. [Google Scholar] [CrossRef] [PubMed]
- Lauritano, C.; Martin, J.; De La Cruz, M.; Reyes, F.; Romano, G.; Ianora, A. First identification of marine diatoms with anti-tuberculosis activity. Sci. Rep. 2018, 8, 2284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.-H.; Chung, T.D.Y.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Ono, K.; Demchak, B.; Ideker, T. Cytoscape tools for the web age: D3. js and Cytoscape. js exporters version 2. F1000Research 2014, 3, 1–13. [Google Scholar] [CrossRef]









| Strain | Geographic Origin * | Ecology | Micromonospora Species | Similarity (%) |
|---|---|---|---|---|
| CA-107814 | Costa Rica (1) | soil-rice cultivation | M. terminaliae | 99.48 |
| CA-108000 | Costa Rica (1) | soil-rice cultivation | M. endophytica | 100 |
| CA-184181 | Mexico (2) | soil | M. siamensis | 99.32 |
| CA-214658 | Spain (3) | marine cave sediment | M. endophytica | 99.63 |
| CA-214671 | Spain (3) | marine cave sediment | M. chaiyaphumensis | 99.84 |
| CA-218877 | Spain (3) | marine invertebrate Porifera sp. | M. endophytica | 100 |
| CA-238377 | Georgia (4.a) | rhizosphere soil of Pteridium tauricum | M. terminaliae | 99.54 |
| CA-238397 | Georgia (4.a) | rhizosphere soil of Pteridium tauricum | M. chalcea | 100 |
| CA-238398 | Georgia (4.a) | rhizosphere soil of Pteridium tauricum | M. siamensis | 99.32 |
| CA-243027 | Georgia (4.a) | rhizosphere soil of Pteridium tauricum | M. terminaliae | 100 |
| CA-243168 | Central African Republic (5) | forest organic humid soil | M. endophytica | 100 |
| CA-244160 | Union of the Comoros (6.a) | forest organic dry soil | M. terminaliae | 99.53 |
| CA-244161 | Union of the Comoros (6.a) | forest organic dry soil | M. terminaliae | 99.47 |
| CA-244669 | Union of the Comoros (6.b) | forest organic humid soil | M. terminaliae | 99.41 |
| CA-244673 | Union of the Comoros (6.b) | forest organic humid soil | M. chalcea | 100 |
| CA-244674 | Union of the Comoros (6.b) | forest organic humid soil | M. endophytica | 99.31 |
| CA-244675 | Union of the Comoros (6.b) | forest organic humid soil | M. terminaliae | 99.49 |
| CA-246501 | Georgia (4.a) | rhizosphere soil of Pteridium tauricum | M. terminaliae | 99.53 |
| CA-246506 | Georgia (4.a) | rhizosphere soil of Pteridium tauricum | M. aurantiaca | 99.64 |
| CA-248285 | New Zealand (7) | swampy soil | M. soli | 99.07 |
| CA-248314 | New Zealand (7) | swampy soil | M. endophytica | 99.66 |
| CA-248649 | Union of the Comoros (6.c) | forest organic soil | M. terminaliae | 100 |
| CA-249271 | Union of the Comoros (6.d) | forest organic soil | M. endophytica | 99.38 |
| CA-249379 | Union of the Comoros (6.a) | forest organic dry soil | M. terminaliae | 99.54 |
| CA-251294 | Union of the Comoros (6.c) | forest organic soil | M. terminaliae | 99.51 |
| CA-253038 | Georgia (4.b) | rhizosphere soil of Populus canescens | M. siamensis | 99.29 |
| CA-259211 | Georgia (4.c) | rhizosphere soil of Ranunculus buhsei | No data | - |
| Compound | m/z Detected, Adduction | Theoretical Mass (Δ ppm) | Molecular Formula | Retention Time |
|---|---|---|---|---|
| spirotetronates | ||||
| phocoenamicin | 1088.4982, [M + NH4]+ | 1088.4980 (+0.2) | C56H75ClO18 | 6.44 |
| phocoenamicin B | 1104.4927, [M + NH4]+ | 1104.4929 (−0.2) | C56H75ClO19 | 5.53 |
| phocoenamicin C | 1104.4933, [M + NH4]+ | 1104.4929 (+0.4) | C56H75ClO19 | 6.02 |
| maklamicin | 542.3478, [M + NH4]+ | 542.3476 (+0.4) | C32H44O6 | 6.38 |
| siderophores | ||||
| nocardamine | 601.3551, [M + H]+ | 601.3556 (−0.8) | C27H48N6O9 | 1.26 |
| deoxynocardamine | 585.3589, [M + H]+ | 585.3606 (−2.9) | C27H48N6O8 | 1.02 |
| demethylenenocardamine | 587.3396, [M + H]+ | 587.3399 (−0.5) | C26H46N6O9 | 0.95 |
| terragine D | 479.2848, [M + H]+ | 479.2864 (−3.3) | C24H38N4O6 | 2.54 |
| terragine B | 319.1652, [M + H]+ | 319.1652 (0.0) | C17H22N2O4 | 2.80 |
| IC 202B | 533.3275, [M + H]+ | 533.3293 (−3.4) | C23H44N6O8 | 1.51 |
| deferoxamine | 561.3604, [M + H]+ | 561.3606 (−0.4) | C25H48N6O8 | 0.70 |
| proferrioxamine A1 | 547.3450, [M + H]+ | 547.3450 (0.0) | C24H46N6O8 | 0.65 |
| desferrioxamine D1 | 603.3695, [M + H]+ | 603.3712 (−1.1) | C27H50N6O9 | 1.06 |
| legonoxamine A | 637.3914, [M + H]+ | 637.3919 (−0.8) | C31H52N6O8 | 2.34 |
| legonoxamine G | 623.3763, [M + H]+ | 623.3763 (0.0) | C30H50N6O8 | 2.21 |
| legonoxamine H | 665.3861, [M + H]+ | 665.3869 (−1.2) | C32H52N6O9 | 2.47 |
| microferrioxamine B | 729.5473, [M + H]+ | 729.5484 (−1.5) | C37H72N6O8 | 4.78 |
| microferrioxamine C | 743.5632, [M + H]+ | 743.5641 (−1.2) | C38H74N6O8 | 4.97 |
| microferrioxamine D | 757.5792, [M + H]+ | 757.5797 (−0.7) | C39H76N6O8 | 5.21 |
| acyl ferrioxamine 2 | 679.4015, [M + H]+ | 679.4025 (−1.4) | C33H54N6O9 | 2.78 |
| various | ||||
| daidzein | 255.0648, [M + H]+ | 255.0652 (−1.6) | C15H10O4 | 2.55 |
| genistein | 271.0594, [M + H]+ | 271.0601 (−2.6) | C15H10O5 | 3.20 |
| glycitein | 285.0754, [M + H]+ | 285.0757 (−1.1) | C16H12O5 | 2.67 |
| antascomicin D | 646.3946, [M + H]+ | 646.3950 (−0.6) | C36H55NO9 | 3.80 |
| 21-demethyl-leptomycin A | 513.3208, [M + H]+ | 513.3211 (−0.6) | C31H44O6 | 5.07 |
| indothiazinone-4-carboxylic acid | 273.0327, [M + H]+ | 273.0328 (−0.4) | C13H8N2O3S | 2.98 |
| anandin A | 358.2743, [M + H]+ | 358.2741 (+0.6) | C23H35NO2 | 4.52 |
| ganefromycin epsilon | 644.3794, [M + H]+ | 644.3793 (+0.2) | C36H53NO9 | 4.70 |
| actiphenol | 276.1238, [M + H]+ | 276.1230 (+2.9) | C15H17NO4 | 2.48 |
| Nb-Acetyltryptamine | 203.1177, [M + H]+ | 203.1179 (−1.0) | C12H14N2O | 2.41 |
| N-Acetyltyramine | 180.1023, [M + H]+ | 180.1019 (+2.2) | C10H13NO2 | 0.92 |
| actiphenamide | 294.1331, [M + H]+ | 294.1336 (−1.7) | C15H19NO5 | 2.48 |
| antibiotic BE 54476 | 392.2429, [M + H]+ | 392.2431 (−0.5) | C22H33NO5 | 4.14 |
| Compound | m/z Detected, Adduction | Theoretical Mass (Δ ppm) | Molecular Formula Predicted | Retention Time |
|---|---|---|---|---|
| spirotetronates | ||||
| phocoenamicin derivative | 1120.4862, [M + NH4]+ | 1120.4878 (−1.4) | C56H75ClO20 | 5.29 |
| phocoenamicin derivative | 1032.4713, [M + NH4]+ | 1032.4718 (−0.5) | C53H71ClO17 | 5.70 |
| phocoenamicin derivative | 1122.4584, [M + NH4]+ | 1122.4590 (−0.5) | C56H74Cl2O18 | 6.71 |
| phocoenamicin derivative | 1136.4795, [M + NH4]+ | 1136.4828 (−2.9) | C56H75ClO21 | 4.54 |
| phocoenamicin derivative | 920.4994, [M + NH4]+ | 920.5002 (−0.9) | C48H70O16 | 5.24 |
| phocoenamicin derivative | 936.4939, [M + NH4]+ | 936.4951 (−1.3) | C48H70O17 | 4.46 |
| phocoenamicin derivative | 644.3787, [M + NH4]+ | 644.3793 (−0.9) | C36H50O9 | 5.55 |
| phocoenamicin derivative | 774.4418, [M + NH4]+ | 744.4423 (−0.7) | C42H60O12 | 5.19 |
| maklamicin derivative | 509.3250, [M + H]+ | 509.3262 (−2.4) | C32H44O5 | 7.30 |
| maklamicin derivative | 495.3091, [M + H]+ | 495.3105 (−2.8) | C31H42O5 | 7.09 |
| maklamicin derivative | 511.3060, [M + H]+ | 511.3054 (+1.2) | C31H42O6 | 6.12 |
| maklamicin derivative | 539.3350, [M + H]+ | 539.3367 (−3.2) | C33H46O6 | 6.60 |
| maklamicin derivative | 523.3049, [M + H]+ | 523.3054 (−1.0) | C32H42O6 | 6.91 |
| siderophores | ||||
| nocardamine derivative | 599.3379, [M + H]+ | 599.3399 (−3.3) | C27H46N6O9 | 1.85 |
| deferoxamine derivative | 631.4384, [M + H]+ | 631.4389 (−0.8) | C30H58N6O8 | 2.88 |
| deferoxamine derivative | 645.4531, [M + H]+ | 645.4545 (−2.2) | C31H60N6O8 | 3.23 |
| deferoxamine derivative | 687.4638, [M + H]+ | 687.4651 (−1.9) | C33H62N6O9 | 3.44 |
| deferoxamine derivative | 659.4342, [M + H]+ | 659.4338 (+0.6) | C31H58N6O9 | 2.94 |
| deferoxamine derivative | 671.4694, [M + H]+ | 671.4702 (−1.0) | C33H62N6O8 | 3.66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokkini, M.; González Heredia, C.; Oves-Costales, D.; de la Cruz, M.; Sánchez, P.; Martín, J.; Vicente, F.; Genilloud, O.; Reyes, F. Exploring Micromonospora as Phocoenamicins Producers. Mar. Drugs 2022, 20, 769. https://doi.org/10.3390/md20120769
Kokkini M, González Heredia C, Oves-Costales D, de la Cruz M, Sánchez P, Martín J, Vicente F, Genilloud O, Reyes F. Exploring Micromonospora as Phocoenamicins Producers. Marine Drugs. 2022; 20(12):769. https://doi.org/10.3390/md20120769
Chicago/Turabian StyleKokkini, Maria, Cristina González Heredia, Daniel Oves-Costales, Mercedes de la Cruz, Pilar Sánchez, Jesús Martín, Francisca Vicente, Olga Genilloud, and Fernando Reyes. 2022. "Exploring Micromonospora as Phocoenamicins Producers" Marine Drugs 20, no. 12: 769. https://doi.org/10.3390/md20120769