Comparison of Transcriptome Differences between Two Rice Cultivars Differing in Cadmium Translocation from Spike-Neck to Grain
Abstract
:1. Introduction
2. Results
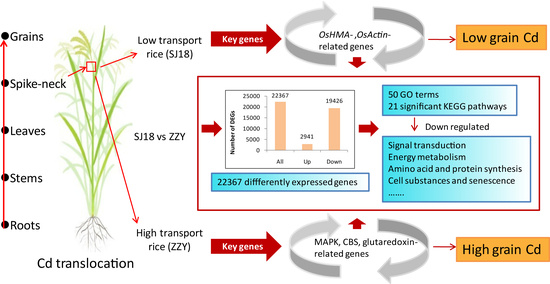
2.1. Cadmium and Its Transfer in Rice Plant
2.2. RNA-Seq Data, qRT-PCR Validation, and Function Annotation
2.3. Differentially Expressed Genes and GO Enrichment
2.4. KEGG Enrichment of Differentially Expressed Genes
2.5. WGCNA of DEGs
3. Discussion
3.1. Down-Regulation of Numerous Physiological Activities May Characterize Low Cd-Transporting Culvitars
3.2. Different Cd Translocation Efficiencies among Varieties Related to Different Core Mechanisms
4. Materials and Methods
4.1. Experiment Site and Soil Properties
4.2. Rice Cultivars and Field Management
4.3. Sampling and Determination of Cd
4.4. Transcriptome Analysis of Spike-Neck Samples
4.5. Quantitative Reverse-Transcription PCR Validation
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hussain, B.; Umer, M.J.; Li, J.; Ma, Y.; Abbas, Y.; Ashraf, M.N.; Tahir, N.; Ullah, A.; Gogoi, N.; Farooq, M. Strategies for reducing cadmium accumulation in rice grains. J. Clean. Prod. 2021, 286, 125557. [Google Scholar] [CrossRef]
- Kulsum, P.S.P.G.; Khanam, R.; Das, S.; Nayak, A.K.; Tack, F.M.G.; Meers, E.; Vithanage, M.; Shahid, M.; Kumar, A.; Chakraborty, S.; et al. A state-of-the-art review on cadmium uptake, toxicity, and tolerance in rice: From physiological response to remediation process. Environ. Res. 2023, 220, 115098. [Google Scholar] [CrossRef] [PubMed]
- Tahir, N.; Ullah, A.; Tahir, A.; Rashid, H.U.; Rehman, T.u.; Danish, S.; Hussain, B.; Akca, H. Strategies for reducing Cd concentration in paddy soil for rice safety. J. Clean. Prod. 2021, 316, 128116. [Google Scholar]
- Wang, L.; Zhang, Q.; Liao, X.; Li, X.; Zheng, S.; Zhao, F. Phytoexclusion of heavy metals using low heavy metal accumulating cultivars: A green technology. J. Hazard. Mater. 2021, 413, 125427. [Google Scholar] [CrossRef] [PubMed]
- Zeb, A.; Liu, W.; Lian, Y.; Zheng, Z.; Meng, L.; Chen, C.; Song, X. Selection and breeding of pollution-safe cultivars (PSCs)—An eco-friendly technology for safe utilization of heavy metal(loid) contaminated soils. Environ. Technol. Inno. 2022, 25, 102142. [Google Scholar] [CrossRef]
- Liu, N.; Huang, X.; Sun, L.; Li, S.; Chen, Y.; Cao, X.; Wang, W.; Dai, J.; Rinnan, R. Screening stably low cadmium and moderately high micronutrients wheat cultivars under three different agricultural environments of China. Chemosphere 2020, 241, 125065. [Google Scholar] [CrossRef] [PubMed]
- Duan, G.; Shao, G.; Tang, Z.; Chen, H.; Wang, B.; Tang, Z.; Yang, Y.; Liu, Y.; Zhao, F.J. Genotypic and Environmental Variations in Grain Cadmium and Arsenic Concentrations Among a Panel of High Yielding Rice Cultivars. Rice 2017, 10, 9. [Google Scholar] [CrossRef]
- Li, X.; Zhou, D. A Meta-Analysis on Phenotypic Variation in Cadmium Accumulation of Rice and Wheat: Implications for Food Cadmium Risk Control. Pedosphere 2019, 29, 545–553. [Google Scholar] [CrossRef]
- Hongjiang, Z.; Xizhou, Z.; Tingxuan, L.; Fu, H. Variation of cadmium uptake, translocation among rice lines and detecting for potential cadmium-safe cultivars. Environ. Earth Sci. 2014, 71, 277–286. [Google Scholar] [CrossRef]
- Yu, H.; Guo, J.; Li, Q.; Zhang, X.; Huang, H.; Huang, F.; Yang, A.; Li, T. Characteristics of cadmium immobilization in the cell wall of root in a cadmium-safe rice line (Oryza sativa L.). Chemosphere 2020, 241, 125095. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Xiao, W.; Chen, D.; Hu, J.; Gao, N.; Huang, M.; Ye, X. Comparative transcriptomic analysis reveals the important process in two rice cultivars with differences in cadmium accumulation. Ecotoxicol. Environ. Saf. 2023, 252, 114629. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yu, H.; Zhang, X.; Ye, D.; Huang, H.; Wang, Y.; Zheng, Z.; Li, T. A transcriptomic view of cadmium retention in roots of cadmium-safe rice line (Oryza sativa L.). J. Hazard. Mater. 2021, 418, 126379. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, H.; Reinfelder, J.R.; Liang, X.; Sun, C.; Liu, C.; Li, F.; Yi, J. A transcriptomic (RNA-seq) analysis of genes responsive to both cadmium and arsenic stress in rice root. Sci. Total Environ. 2019, 666, 445–460. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Liu, J.; Wu, D.; Liu, Y.; Cen, W.; Wang, S.; Li, R.; Luo, J. Weighted gene coexpression network analysis-based identification of key modules and hub genes associated with drought sensitivity in rice. BMC Plant Biol. 2020, 20, 478. [Google Scholar]
- Zeng, Z.; Zhang, S.; Li, W.; Chen, B.; Li, W. Gene-coexpression network analysis identifies specific modules and hub genes related to cold stress in rice. BMC Genom. 2022, 23, 251. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Xie, H.; Wei, X.; Dossa, K.; Yu, Y.; Hui, S.; Tang, G.; Zeng, X.; Yu, Y.; Hu, P.; et al. WGCNA Analysis of Salt-Responsive Core Transcriptome Identifies Novel Hub Genes in Rice. Genes 2019, 10, 719. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Xiao, W.; Chen, D.; Hu, J.; Gao, N.; Huang, M.; Ye, X. Comparative transcriptome analysis reveals key genes and coordinated mechanisms in two rice cultivars differing in cadmium accumulation. Chemosphere 2023, 338, 139489. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, K.; Zhang, X.; Huang, H.; Huang, F.; Zhang, L.; Wang, Y.; Li, T.; Yu, H. Genetic properties of cadmium translocation from straw to brown rice in low-grain cadmium rice (Oryza sativa L.) line. Ecotoxicol. Environ. Saf. 2019, 182, 109422. [Google Scholar] [CrossRef]
- Tan, M.; Cheng, D.; Yang, Y.; Zhang, G.; Qin, M.; Chen, J.; Chen, Y.; Jiang, M. Co-expression network analysis of the transcriptomes of rice roots exposed to various cadmium stresses reveals universal cadmium-responsive genes. BMC Plant Biol. 2017, 17, 194. [Google Scholar] [CrossRef]
- Xue, D.; Jiang, H.; Deng, X.; Zhang, X.; Wang, H.; Xu, X.; Hu, J.; Zeng, D.; Guo, L.; Qian, Q. Comparative proteomic analysis provides new insights into cadmium accumulation in rice grain under cadmium stress. J. Hazard. Mater. 2014, 280, 269–278. [Google Scholar] [CrossRef]
- Zeng, T.; Fang, B.; Huang, F.; Dai, L.; Tang, Z.; Tian, J.; Cao, G.; Meng, X.; Liu, Y.; Lei, B.; et al. Mass spectrometry-based metabolomics investigation on two different indica rice grains (Oryza sativa L.) under cadmium stress. Food Chem. 2021, 343, 128472. [Google Scholar] [CrossRef] [PubMed]
- Mwamba, T.M.; Islam, F.; Ali, B.; Lwalaba, J.L.W.; Gill, R.A.; Zhang, F.; Farooq, M.A.; Ali, S.; Ulhassan, Z.; Huang, Q.; et al. Comparative metabolomic responses of low- and high-cadmium accumulating genotypes reveal the cadmium adaptive mechanism in Brassica napus. Chemosphere 2020, 250, 126308. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Xie, Y.; He, Z.; Zhang, J.; Tang, Y.; Zhou, X. Network response of two cherry tomato (Lycopersicon esculentum) cultivars to Cadmium stress as revealed by transcriptome analysis. Ecotoxicol. Environ. Saf. 2021, 222, 112473. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Yang, Y.; Yang, Z. Molecular dissection of cadmium-responsive transcriptome profile in a low-cadmium-accumulating cultivar of Brassica parachinensis. Ecotoxicol. Environ. Saf. 2019, 176, 85–94. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, M.; Tao, Q.; Luo, J.; Li, J.; Guo, X.; Liang, Y.; Yang, X.; Li, T. A comparative study of root cadmium radial transport in seedlings of two wheat (Triticum aestivum L.) genotypes differing in grain cadmium accumulation. Environ. Pollut. 2020, 266, 115235. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Roychoudhury, A. Transcriptome Profiling of Abiotic Stress-Responsive Genes During Cadmium Chloride-Mediated Stress in Two Indica Rice Varieties. J. Plant Growth Regul. 2017, 37, 657–667. [Google Scholar] [CrossRef]
- Lu, C.; Zhang, L.; Tang, Z.; Huang, X.Y.; Ma, J.F.; Zhao, F.J. Producing cadmium-free Indica rice by overexpressing OsHMA3. Environ. Int. 2019, 126, 619–626. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Ma, J.F. Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 2014, 65, 6013–6021. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Gao, E.; Hussey, P.J. Autophagosome Biogenesis in Plants: An Actin Cytoskeleton Perspective. Trends Plant Sci. 2020, 25, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Ding, Y.; Jiang, Q.; Wang, F.; Sun, J.; Zhu, C. The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 2017, 36, 235–242. [Google Scholar] [CrossRef]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Opdenakker, K.; Remans, T.; Vangronsveld, J.; Cuypers, A. Mitogen-Activated Protein (MAP) Kinases in Plant Metal Stress: Regulation and Responses in Comparison to Other Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2012, 13, 7828–7853. [Google Scholar] [CrossRef] [PubMed]
- Šamajová, O.; Plíhal, O.; Al-Yousif, M.; Hirt, H.; Šamaj, J. Improvement of stress tolerance in plants by genetic manipulation of mitogen-activated protein kinases. Biotechnol. Adv. 2013, 31, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Opdenakker, K.; Remans, T.; Keunen, E.; Vangronsveld, J.; Cuypers, A. Exposure of Arabidopsis thaliana to Cd or Cu excess leads to oxidative stress mediated alterations in MAPKinase transcript levels. Environ. Exp. Bot. 2012, 83, 53–61. [Google Scholar] [CrossRef]
- Yeh, C.M.; Chien, P.S.; Huang, H.J. Distinct signalling pathways for induction of MAP kinase activities by cadmium and copper in rice roots. J. Exp. Bot. 2006, 58, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Yang, Y. Disease Resistance and Abiotic Stress Tolerance in Rice Are Inversely Modulated by an Abscisic Acid–Inducible Mitogen-Activated Protein Kinase. Plant Cell 2003, 15, 745–759. [Google Scholar] [CrossRef]
- Wu, Q.; Yang, J.; Cheng, N.; Hirschi, K.D.; White, F.F.; Park, S. Glutaredoxins in plant development, abiotic stress response, and iron homeostasis: From model organisms to crops. Environ. Exp. Bot. 2017, 139, 91–98. [Google Scholar] [CrossRef]
- Ali, F.; Li, Y.; Li, F.; Wang, Z. Genome-wide characterization and expression analysis of cystathionine β-synthase genes in plant development and abiotic stresses of cotton (Gossypium spp.). Int. J. Biol. Macromol. 2021, 193, 823–837. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, R.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Overexpression of Rice CBS Domain Containing Protein Improves Salinity, Oxidative, and Heavy Metal Tolerance in Transgenic Tobacco. Mol. Biotechnol. 2012, 52, 205–216. [Google Scholar] [CrossRef]
- Dörmanna, P.; Gopalan, S.; He, S.Y.; Benning, C. A gene family in Arabidopsis thaliana with sequence similarity to NDR1 and HIN1. Plant Physiol. Bioch. 2000, 38, 789–796. [Google Scholar] [CrossRef]
- Ding, X.; Hou, X.; Xie, K.; Xiong, L. Genome-wide identification of BURP domain-containing genes in rice reveals a gene family with diverse structures and responses to abiotic stresses. Planta 2009, 230, 149–163. [Google Scholar] [CrossRef]
- Javed, T.; Gao, S.-J. WRKY transcription factors in plant defense. Trends Genet. 2023, 39, 787–801. [Google Scholar] [CrossRef]
- Tian, W.; Huang, Y.; Li, D.; Meng, L.; He, T.; He, G. Identification of StAP2/ERF genes of potato (Solanum tuberosum) and their multiple functions in detoxification and accumulation of cadmium in yest: Implication for Genetic-based phytoremediation. Sci. Total Environ. 2022, 810, 152322. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Zhang, Q.; Xiao, W.; Chen, D.; Hu, J.; Gao, N.; Huang, M.; Ye, X. Comparison of Transcriptome Differences between Two Rice Cultivars Differing in Cadmium Translocation from Spike-Neck to Grain. Int. J. Mol. Sci. 2024, 25, 3592. https://doi.org/10.3390/ijms25073592
Zhao S, Zhang Q, Xiao W, Chen D, Hu J, Gao N, Huang M, Ye X. Comparison of Transcriptome Differences between Two Rice Cultivars Differing in Cadmium Translocation from Spike-Neck to Grain. International Journal of Molecular Sciences. 2024; 25(7):3592. https://doi.org/10.3390/ijms25073592
Chicago/Turabian StyleZhao, Shouping, Qi Zhang, Wendan Xiao, De Chen, Jing Hu, Na Gao, Miaojie Huang, and Xuezhu Ye. 2024. "Comparison of Transcriptome Differences between Two Rice Cultivars Differing in Cadmium Translocation from Spike-Neck to Grain" International Journal of Molecular Sciences 25, no. 7: 3592. https://doi.org/10.3390/ijms25073592