1. Introduction
The potential application of MSCs in regenerative medicine was initially focused on cell transplantation into the patients and the subsequent differentiation of those MSCs into a specific (resident) cell type to replace lost or damaged tissue. However, the different steps of cell manipulation required prior to the implantation of those cells into a given tissue has detrimental effects on cell viability. The need to expand in vitro the MSCs cells prior to implantation, to achieve a cell number sufficient to undergo many of the procedures, also increases genomic instability having a negative effect on the regeneration potential of those cells. Moreover, the administration route, such as local or systemic injection, can exert important mechanical stress on the cells, also limiting their viability.
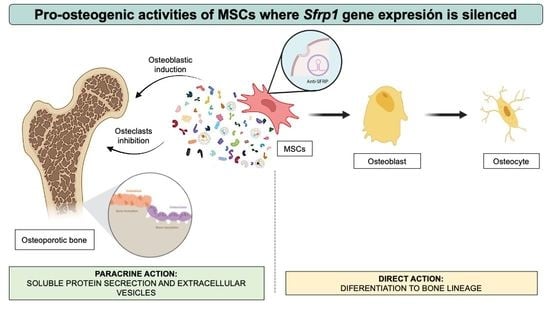
In addition, current evidence in animal models shows that the beneficial effects of MSCs in regenerative therapies are achieved primarily through their important paracrine action on the bone marrow (BM) microenvironment rather than through their ability to differentiate into various cell lineages [
1,
2]. Bone marrow MSCs (BM-MSCs) secrete a range of bioactive products into the surrounding microenvironment, which have trophic effects on neighboring cells stimulating numerous biological processes [
3,
4]. The soluble factors and extracellular vesicles (EVs) produced by BM-MSCs are considered the main modulators of cellular crosstalk at the BM microenvironment, greatly influencing bone homeostasis and enhancing bone repair and regeneration [
5,
6]. These findings prompted a change in paradigm, with the researchers focusing their attention on the secretome of MSCs as a key tool for MSCs-mediated regenerative medicine.
Although the secretome from MSCs has broad proangiogenic, antifibrotic, anti-inflammatory, and antiapoptotic properties, its plasticity allows us to engineer the composition of the soluble fraction as well as that of the exosomal cargo to produce a tailor-made product that would fit specific therapeutic requirements [
7]. Different approaches have been developed to engineer an MSCs-derived secretome or exosomal cargo for improving bone regeneration, from pre-conditioning MSCs with different biochemical compounds, small molecules or cytokines, to subjecting the MSCs to diverse biophysical cues or to directly altering MSCs gene expression through genetic manipulation.
Recently, our team has been developing a method for the systemic management of osteoporosis based on the silencing of inhibitors of the main osteogenic signaling pathways in MSCs [
8,
9]. Silencing of target genes is achieved by using a particular type of Lock Nucleic Acid Antisense Oligonucleotides, known as GapmeRs which, in our system, are encapsulated in hybrid nanoparticles [
8]. These nanoparticles are then delivered to the endogenous MSCs in the BM of a murine osteoporotic model, using an aptamer specifically designed to target these cells [
10]. Although the use of the GapmeRs produces only a transient silencing of the target genes, this seems to be enough to prime MSCs towards osteogenic differentiation. In fact, using this method, which transiently silences
Sfrp1, an antagonist of the Wnt/β-catenin signaling pathway, at the level of the endogenous bone marrow MSCs results in a significant increase in bone mineral density in an osteoporotic mouse model [
10].
In the same way that MSCs potential to differentiate into osteoblasts can be increased by the transient silencing of anti-osteogenic genes, it is also possible that these modifications can alter their secretory profile to change the production of different factors, generating a secretome that has an overall positive effect on all the cells in the BM microenvironment. Hypothetically, the priming process, consisting of the transient silencing of key osteogenic inhibitors, would encompass important changes in gene expression and protein production. The secretome of the primed cells would likely include osteogenic proteins that are synthesized in the cytoplasm and later released outside the cell. Similarly, during their biogenesis, EVs such as exosomes, key constituents of the secretome, would capture and encapsulate cytoplasmic components, including specific proteins and RNA molecules linked to osteogenic differentiation, overall leading to the production of a secretome enriched in osteogenic factors. This secretome could have important paracrine effects on neighboring cells, including those responsible for bone homeostasis. That is, a cell primed for osteogenic differentiation would theoretically produce a more osteogenic secretome that could have similar osteo-regenerative properties than the modified cell.
In this current work, we set out to investigate the effects of the secretome of MSCs where Sfrp1 has been transiently silenced on other cells resident in the BM with a key role in bone homeostasis and further analyze the protein composition of the soluble and vesicular fractions of this secretome to identify the molecules with putative osteo-regenerative properties.
Although there are still limitations in MSCs-derived secretome-based therapies, various methods to engineer secretomes with increased osteo-regenerative potential could open a promising field in bone regeneration, which can attract further investigations.
3. Discussion
Many works have shown that the number of transplanted MSCs that can contribute to tissue regeneration is minimal. Due to their low engraftment rates, it seems now clear that the therapeutic effect of MSCs is not only achieved by promoting the osteogenic differentiation of other MSCs [
11,
12] but most of the regenerative effects observed after MSCs implantation can be attributed to their paracrine action [
5]. These results prompted a change in paradigm. In view of this, researchers have focused their attention on the secretome of MSCs as a key tool for MSCs-mediated regenerative medicine [
13].
The bone microenvironment is complex, harboring many different cells that form an intricate communication network [
14]. Even when focusing exclusively on the interactions related to bone homeostasis and remodeling, with the simplified osteocyte–osteoclast–osteoblast axis, external influences play a significant role, as they can modify the interactions within the axis, as seen in postmenopausal osteoporosis [
15].
Here, we set out to investigate how the paracrine action of MSCs where
Sfrp1 has been silenced contributes to the increase in bone regeneration seen in an osteoporotic murine mouse model [
10].
Regarding the analysis of the effect of the mCM-Sfrp1 on osteoblasts, our experiments indicate a positive effect on osteoblast differentiation in vitro in the C3H19T1/4 murine cell line. However, the results using human primary osteoporotic MSCs were not as clear. We did observe a tendency to a reduction in the RUNX2 expression levels in the presence of the hCM-SFRP1, which will be indicative of an accelerated osteogenic differentiation, as well as a trend to the increase of the late osteogenic marker BGLAP, although these data were not significant. This might be due to the small number of samples analyzed, which is directly related to the scarcity of samples that fit our stringent patient selection criteria in terms of sex, age, and medical history. The increase in the number of these samples would probably allow drawing more clear conclusions. However, the fact that we detected an increase in the levels of mineralized tissue formation in vitro when these cells are treated with the hCM-SFRP1 would certainly suggest a positive effect on osteogenic differentiation. Although our in vitro results using the C3H10T1/2 cell line do point to a positive effect of the mCM-Sfrp1 on C3H10T1/2 differentiation, we did not observe a significant effect of the mCM-Sfrp1 over the analyzed osteogenic markers ex vivo. This might be explained by the relatively low presence of MSCs and osteoblasts in the calvaria bone discs relative to the presence of osteocytes, which are, by far, the most abundant cells in this kind of samples.
Osteocytes impact bone remodeling by secreting multiple factors that drive the differentiation and activity of other bone cells, mainly through the secretion of RANKL and OPG [
16]. By expressing and secreting RANKL, osteocytes regulate the differentiation of the OCs precursors, which belong to the hematopoietic lineage [
17]. OPG, however, acts as an antagonist to RANKL, inhibiting osteoclast differentiation by competing with RANKL for the binding to the RANK receptor. Therefore, the ratio between the expression of RANKL and OPG can be used as a marker of bone turnover. In our case, we did not detect a significant reduction of the RANKL/OPG ratio after incubating the osteocytic cell line MLO-A5 with the CM where
Sfrp1 was silenced (mCM-
Sfrp1), but did observe a highly significant reduction of this ratio in the more complex microenvironment of an ex vivo culture of calvaria bone discs, when this has been treated with the mCM-
Sfrp1 compared to mCM-Ctrl. This suggests that this CM-
Sfrp1 might act on the osteocytes, modulating the production of RANKL and OPG in favor of the latter, therefore potentially contributing to decrease bone resorption previously observed in our osteoporotic murine models [
10].
Osteocytes also act as regulators of their own precursors, the osteoblast. The most important factor in this regard is sclerostin, encoded by the
Sost gene. Sclerostin, almost exclusively produced by osteocytes, is an important inhibitor of osteoblast activity and differentiation, as it blocks the Wnt/β-catenin signaling pathway by binding to the LRP4/5/6 membrane receptors. Similarly, DKK1, also secreted by osteocytes, competes with Wnt proteins for the binding to the LRP4/5/6 membrane receptors also inhibiting Wnt signaling [
18]. Our in vitro analysis on the osteocytic cell line MLO-A5 showed that the treatment with the CM-
Sfrp1 does not lead to significant decrease in the expression of the Wnt pathway inhibitor sclerostin. However, the ex vivo culture showed an opposite effect, observing a trend to an increase of
Sost expression, although this was again not significant. This variation between the in vitro and ex vivo experiments could be explained by the more complex microenvironment present in the ex vivo culture, where the interactions among different cells present in the bone sample interfere with the single effect of the CM on the osteocytes. Interestingly,
Dkk1 expression was also showed to be downregulated in the bone culture. This would indicate that the downregulation of
Dkk1 in osteocytes, in contact with the CM-
Sfrp1, potentially contributes to the creation of a pro-osteogenic microenvironment.
The effect of the CM on osteoclasts was also analyzed in the ex vivo culture. The analysis of the expression of osteoclastic markers showed a tendency towards an inhibitory effect. The expression of the main osteoclastic transcriptional factor Nfatc1 was significantly reduced in both CM, and thus, this could not be linked to a specific effect of Sfrp1 silencing on secretome composition. The expression of Ctsk also presented a tendency to decrease in both experimental groups. Interestingly, the expression of Mmp9, a proteolytic enzyme with a key role in bone resorption highly expressed in osteoclasts, was significantly downregulated in samples pre-treated with the mCM-Sfpr1 suggesting a negative effect of the mCM-Sfrp1 on the bone resorption process.
To further elucidate the mechanisms involved in the pro-regenerative effects of the secretome of C3H10T1/2 cells
Sfrp1 was silenced both the protein soluble fraction (SF) and protein present in the exosomal cargo were analyzed through mass spectrometry. Among the proteins significantly more abundant in the cargo of exosomes isolated from the mCM-
Sfrp1 (Exosomal fraction_EF), we found Segment Polarity Protein Dishvelled Homolog DVL-1 (DVL1). This result is not totally unexpected, as the silencing of
Sfrp1 and thus the sustained activation of the Wnt/β-catenin signaling pathway could certainly promote the expression of DVL1, since this protein is a key component of the Wnt/β-catenin signaling pathway that transduces the Wnt signal to downstream effector molecules [
19]. Due to the mechanistic of the exosome biogenesis process, the higher presence of this protein in the cargo of the exosomes produced by cells where
Sfrp1 has been silenced would likely reflect a higher concentration of this protein in the cytoplasm of cells with a high activity of the Wnt/β-catenin signaling pathway and, thus, a higher osteogenic potential. Regarding the soluble fraction of this secretome, one of the proteins found to be significantly decreased in the soluble fraction of mCM-
Sfrp1 is the Matrix Gla protein (MGP). There is some discrepancy between the studies published up to this date regarding the role of this protein in bone regeneration. Some authors have shown that MGP has a role in promoting osteoblasts proliferation, differentiation, and mineralization [
20]. However, our results would be at odds with these findings and in agreement with other works that point to MGP as a potent inhibitor of calcification. MGP would act on sequestering BMP, in particular BMP2, in vitro, therefore inhibiting osteogenic differentiation [
21]. Interestingly, another protein also over-represented in the soluble fraction of the mCM-
Sfrp1 is CCN4. This protein characteristic of the bone extracellular matrix belongs to the CCN family, named from its founding members (Cyr61 CTGF, Nov, 6). CCN4 is typically found expressed in newly formed bone [
22,
23] and is found upregulated in healing bone after induced fracture, highlighting its key role in skeletal homeostasis [
24]. Importantly, this protein is able to promote osteogenic differentiation in osteoprogenitor cells cultured in vitro when added exogenously [
24,
25]. Therefore, the significant enrichment of this protein in the secretome of MSCs where
Sfrp1 has been silencing could have a key role in promoting bone regeneration and might explain, at least in part, the effects seen in vitro in the C3H10T1/2 cells when exposed to the mCM-
Sfrp1.
Overall, the paracrine effect of endogenous MSCs on other cells in the bone marrow microenvironment, mainly on the osteocytes, after the inhibition of the Wnt antagonist Sfrp1 in those cells, would overall enhance bone regeneration. This paracrine effect would add up to the already positive effect directly exerted by the silencing of Sfrp1 in those MSCs, which would trigger their osteogenic differentiation. Therefore, the secretome of the modified cells would provide an added benefit to potential bone regeneration treatments. Our results suggest that the treated MSCs would be able to create a microenvironment that is not only pro-osteogenic, since it will promote MSCs differentiation to the osteoblastic lineage, but also anti-resorptive, by inhibiting osteoclast differentiation and reducing the available osteocyte-derived RANKL.
The use of this pro-osteogenic secretome in cell-free therapies would allow us to bypass the need of directly silencing key regulatory genes at the endogenous MSCs by using LNA-ASOs, thus hypothetically reducing the potential side effects associated to the use of this molecules [
26,
27].
4. Materials and Methods
4.1. Cell Culture and Osteogenic Differentiation
Three different immortalized cell lines were used in this work. The murine MSC line C3H10T1/2 (Clone 8, Ref. CCL-226, ATCC, Manassas, VA, USA) and the human MSC line ASC52telo (Ref. SCRC4000, ATCC, Manassas, VA, USA) were cultured in Dulbecco’s Modified Eagle’s Medium, DMEM (Gibco Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% FBS and 1% penicillin–streptomycin. In addition, ASC57telo needed supplementation with 0.2 mg/mL geneticin (G418 Sulfate, Corning, Manassas, VA, USA). Murine osteocyte cell line MLO-A5 [
28] needed α-MEM culture media (Gibco Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 2.5% FBS, 2.5% BCS (GE Healthcare Life Science, Logan, UT, USA), 1% penicillin–streptomycin,). For the MLO-A5 cells as these are non-adherent cells, all flasks and plates were coated with rat tail collagen.
For osteogenic differentiation of C3H10T1/2, 25.000 cells/cm2 were seeded and incubated overnight to allow attachment. Osteogenic induction was achieved 24h later by replacing the culture media with osteogenic media. Osteogenic media consist of DMEM supplemented with 20 mM β-glycerophosphate, 50 µM ascorbic acid, and 1 µM dexamethasone. For hMSCs-OP, osteogenic differentiation was performed in the same manner but using 20,000 cells/cm2.
4.2. GapmeR Delivery
For gene silencing, we used Locked Nucleic Acid Antisense Oligonucleotides (LNA-ASOs) from Exiqon (Qiagen, Venlo, The Netherlands). Specifically, we worked with GapmeRs, a particular type of LNA-ASOs with more resistance to degradation by endonucleases. For initial GapmeR delivery efficiency testing, we used Antisense LNA GapmeR Negative Control A (Ref. 339515 LG00000002-DDA) labeled with fluorescein (FAM) for flow cytometry detection. In addition, we also acquired specific GapmeRs for Sfrp1 silencing in mouse (Ref. 339511 LG00219849-DDA) and human (Ref. 339511 LG00219859-DDA) cells.
The transfection protocol for C3H10T1/2 has been previously published [
8]. Also, efficiency of
Sfrp1 silencing after C3H10T1/2 mouse cell line transfection with the indicated GapmeRs was verified in a previous work [
9]. A similar transfection process was followed for the ASC57telo human cell line transfection. Briefly, 24 h after the C3H10T1/2 cell line was seeded at 12,500 cells/cm
2 and ASC52telo at 15,000 cells/cm
2, culture media was replaced by serum-deprived Opti-MEM I media (Invitrogen, Waltham, MA, USA). OptiMEM I media was mixed with the recommended amount of Dharmafect lipofection agent (Dharmacon, Horizon Discovery, Cambridge, UK). Separately, each GapmeR was also mixed with OptiMEM I media and incubated 5 min at room temperature (RT) prior to dropwise addition over Dharmafect mix. The final resulted mix was incubated for 20 min at RT and, subsequently, added dropwise to the seeded cells. Cells were incubated overnight at 37 °C. One volume of DMEM with 10% FBS and 1% penicillin–streptomycin was added to the wells and incubated at 37 °C for another 24 h.
4.3. Isolation of Human Mesenchymal Stem Cells
Human MSCs (hMSCs) were isolated from femoral heads of female patients (ages from 77, 78, 80, 85, 87, and 89 years old) who suffer from osteoporotic fracture and needed hip replacement surgery. Cell isolation was carried out following previously described protocols [
29]. These cells were cultured in MesenPRO RS Media (Thermo Fisher Scientific, Waltham, MA, USA) enriched with Glutamax and MesenPRO supplements and were left to form colonies and proliferate for 10–14 days at 37 °C and 5% CO
2. Then, hMSCs were expanded to achieve a cell number of 7–10 × 10
5. All patients gave informed consent, and the study protocol was approved by the Comité de Ética en Investigación Clínica de Cantabria.
4.4. Conditioned Media Production
To test the effect of the treated MSCs over other murine bone cells, conditioned media (CM) derived from MSCs previously transfected with specific GapmeRs was produced. For these, 12 million cells seeded in a 100 mm culture plate were transfected using non-specific GapmeR Ctrl and a GapmeR specific for Sfrp1. Forty-eight hours after transfection, the culture media was discarded, and after washing the cells once with PBS, it was replaced with a total of 12 mL of DMEM without FBS. Cells were then incubated for 48 h under normoxic conditions at 37 °C.
Once the incubation was completed, the CM was collected and centrifuged at 400× g for 10′ at 4 °C, followed by another centrifugation of the supernatant at 1000 g for 10′ at 4 °C. Finally, the supernatant was collected and filtered through a 0.22 µm-syringe filter. The supernatant was aliquoted to avoid repeated freezing–thaw cycles and stored at −20 °C until needed.
To test the effect of CM-
Sfrp1 on MSCs from osteoporotic patients, the human cell line ASC57Telo was used. The CM production and collection is identical to that previously described for the murine cell line C3H10t1/2. The transfection method for these cells is described in
Section 4.2.
4.5. Alizarin Red Quantification
Alizarin Red staining quantification was performed by adapting a previously described protocol [
30]. Briefly, each well (24-well plate) was incubated for 30 min with 200 µL of 10% acetic acid with slight shaking. After the incubation, the plates were scraped and the whole volume together with the pellet was transferred into an Eppendorf tube. Tubes were then vortexed for 30 s followed by a 10-min incubation at 85 °C. Tubes were left to cool for 5 min on ice and then centrifuged at RT for 30 min at 13,000 rpm. Once the centrifugation was finished, a total of 150 µL of the supernatant was transferred into a new tube where 57 µL of 10N NaOH was added to each tube. Samples were then vortexed, and each tube was then plated into 2 wells of a 96-well-plate, 100 µL per well. Finally, the plate absorbance was measured at 405 nm.
4.6. Alkaline Phosphatase Activity Quantification
In order to quantify the activity of the alkaline phosphatase enzyme activity, at the end point of the osteogenic differentiation, wells were washed once with PBS and incubated at RT for 5′ with 200 µL of Triton-X100 0.05% to allow cell lysis. Wells were then scraped, and each well content was transferred into an Eppendorf tube. Samples were subjected to 3 cycles of 30 s of sonication, while immersed in ice. Samples were then stored at −80 °C until further analysis.
Enzyme activity quantification was performed by using P-nitrophenol as substrate following previously described methods [
31].
4.7. Analysis of Osteogenic Genes Expression
mRNA was extracted from cell cultures and converted to cDNA as previously described (9) to perform gene expression analysis by real-time qPCR. These analyses were carried out using Taqman assays (Thermo Fisher Scientific, Waltham, MA, USA). For the mouse assays probes, references were as follows: Gapdh Mm99999915-g1, Sfrp1 Mm00489161_m1, Runx2 Mm00501578_m1, Alpl Mm01187117_m1, Bglap Mm03413726_m1. For the human assay probes, references were as follows: GAPDH Hs99999905_m1, SFRP1 Hs00610060_m1, RUNX2 Hs00231692_m1, ALPL Hs00758162_m1, BGLAP Hs01587814_g1.
4.8. Ex Vivo Bone Culture
To test the effect of the CM on the bone, ex vivo cultures of calvaria disks were isolated and cultured following a previously described protocol [
32]. Briefly, calvaria bones were dissected from 12-week-old CD1 mice and cleaned of soft tissues. Two 5 mm bone disks were obtained from each calvaria using a biopsy punch (Ref. 22617, Kai Industries, Seki, Gifu, Japan). Bone disks were transferred into a 96-well plate, and 200 µL of media was added to each well. Bone discs were maintained in culture with 50% CM, both CM-Sfrp1 and CM-Ctrl for 11 days. After this time, bone tissue was disaggregated, and the mRNA was extracted in order to quantify the expression of different bone markers (
Figure 3a). Different conditions of CM were tested, including: a control group without CM, a second group with 50% of mCM-Ctrl, and a third group with 50% mCM-
Sfrp1.Ex vivo culture was maintained for 11 days, and every 2–3 days half of the culture media was changed, maintaining the corresponding media of each condition. Bone disks were washed once with PBS and placed in 5 mL round bottom tubes containing 1 mL of TRIzol. The tissue was disaggregated with a T25 digital ULTRA-TURRAX (Ref. 0003725000, IKA-Werke, Staufen, Germany) dispersing instrument, with an S25N-8G (Ref. 0001024200, IKA-Werke, Staufen, Germany) dispersion tool. During tissue disaggregation, samples were kept on ice, and two disaggregation pulses of 20 s each were performed on each sample to avoid overheating.
After disaggregation, the tubes were centrifuged at 2000 rpm for 5 min, and the TRIzol supernatant was collected. TRIzol was used for RNA extraction, followed by retrotranscription into cDNA and finally for gene expression quantification.
4.9. Secretome Analysis by Mass Spectrometry
To generate the conditioned media (CM), we used 2 × 10
6 C3H10T1/2 cells that had been previously transfected with the Ctrl or the
Sfrp1 GapmeRs. Forty-eight hours after, the transfection media was replaced by DMEM without serum and CM production was allowed for 48 h. Conditioned medium was then collected and centrifuged 3 min at 1000 rpm at 4 °C, followed by second centrifugation at 3000 rpm for 10 min at 4 °C. Exosomal fraction was isolated using ultracentrifugation, 100,000 rpm 1 h at 4 °C in a Beckman ultracentrifuge (Optima L90-k). The supernatant was labelled as a soluble fraction and the pellet as exosomal fraction. The soluble fraction was concentrated using Amicon Ultra 3 KDa centrifugal filters. Both fractions were processed for mass spectrometry analysis using in solution digest protocol. In brief, proteins were denaturalized using 6 M Guanidine Hydrochloride, reduced and alkylate with 5 mM TCEP (tris(2-carboxyethyl)phosphine) and 10 mM Chloroacetamide, and sequentially digested with MS grade Lys-C and trypsin. Peptides were desalted and purified as previously described [
33]. The eluted peptides were lyophilized in a Concentrator Plus (Eppendorf), resuspended in 0.1% TFA and analyzed by LC-MS/MS on a Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer [
34]. Protein identification and quantification was performed using the DIA-N/N software [
35] and the Perseus software after values normalization by total protein in the cell culture. Statistical changes were determined using an adjusted
t-test against control cells with a
p-value 0.05 and a fold change of 2 or higher.