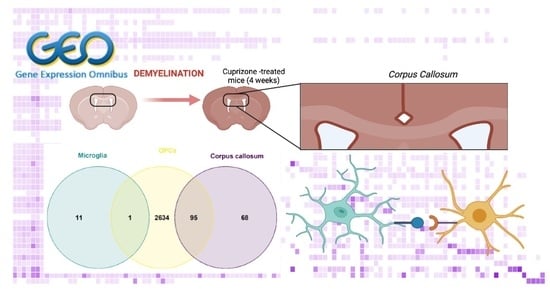
Deciphering the Genetic Crosstalk between Microglia and Oligodendrocyte Precursor Cells during Demyelination and Remyelination Using Transcriptomic Data
Abstract
:1. Introduction
2. Results
2.1. Identification of Demyelination and Remyelination Marker Genes in the Cuprizone Model
2.1.1. Differentially Expressed Genes (DEGs) in the CC
2.1.2. DEGs in Microglia
2.1.3. DEGs in OPCs
2.1.4. Common DEGs in the CC, Microglia and OPCs
2.2. Identification of Common Demyelination Marker Genes in Multiple Sclerosis Samples and in Samples from the Cuprizone Mice Model
2.3. Identification of Remyelination Marker Genes in the CC in the Cuprizone Model
2.4. Cell Type Composition in CC Samples
2.5. Microglia–Oligodendrocyte Interaction in Cuprizone-Induced Demyelination–Remyelination: Potential Receptor–Ligand Pairs to Promote Remyelination
3. Discussion
4. Materials and Methods
4.1. Data
4.2. Identification of Demyelination/Remyelination Biomarkers
4.3. Cell Type Analysis in CC Samples
4.4. Microglia and OPC Crosstalk
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Walton, C.; King, R.; Rechtman, L.; Kaye, W.; Leray, E.; Marrie, R.A.; Robertson, N.; La Rocca, N.; Uitdehaag, B.; van der Mei, I.; et al. Rising prevalence of multiple sclerosis worldwide: Insights from the Atlas of MS, third edition. Mult. Scler. J. 2020, 26, 1816–1821. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, N.; Razavi, S.; Nikzad, E. Multiple Sclerosis: Pathogenesis, Symptoms, Diagnoses and Cell-Based Therapy. Cell J. 2017, 19, 1–10. [Google Scholar]
- Gelfand, J.M. Multiple sclerosis: Diagnosis, differential diagnosis, and clinical presentation. Handb. Clin. Neurol. 2014, 122, 269–290. [Google Scholar] [CrossRef]
- Hauser, S.L.; Cree, B.A. Treatment of Multiple Sclerosis: A Review. Am. J. Med. 2020, 133, 1380–1390.e2. [Google Scholar] [CrossRef] [PubMed]
- Dobson, R.; Giovannoni, G. Multiple sclerosis—A review. Eur. J. Neurol. 2019, 26, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.; Cai, Z.; Rhodes, P.G. Effects of lipopolysaccharide on oligodendrocyte progenitor cells are mediated by astrocytes and microglia. J. Neurosci. Res. 2000, 62, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xu, D.; Wang, S.; Chen, Y.; Li, Z.; Gao, X.; Jiang, L.; Tang, Y.; Peng, Y. Astrocytes induce proliferation of oligodendrocyte progenitor cells via connexin 47-mediated activation of the ERK/Id4 pathway. Cell Cycle 2017, 16, 714–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willis, C.M.; Nicaise, A.M.; Bongarzone, E.R.; Givogri, M.; Reiter, C.R.; Heintz, O.; Jellison, E.R.; Sutter, P.A.; TeHennepe, G.; Ananda, G.; et al. Astrocyte Support for Oligodendrocyte Differentiation can be Conveyed via Extracellular Vesicles but Diminishes with Age. Sci. Rep. 2020, 10, 828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnädelbach, O.; Fawcett, J. Astrocyte influences on oligodendrocyte progenitor migration. Prog. Brain Res. 2001, 132, 97–102. [Google Scholar] [CrossRef]
- Kalafatakis, I.; Karagogeos, D. Oligodendrocytes and Microglia: Key Players in Myelin Development, Damage and Repair. Biomolecules 2021, 11, 1058. [Google Scholar] [CrossRef]
- Santos, E.N.; Fields, R.D. Regulation of myelination by microglia. Sci. Adv. 2021, 7, eabk1131. [Google Scholar] [CrossRef] [PubMed]
- Miron, V.E.; Boyd, A.; Zhao, J.-W.; Yuen, T.J.; Ruckh, J.M.; Shadrach, J.L.; van Wijngaarden, P.; Wagers, A.J.; Williams, A.; Franklin, R.J.M.; et al. M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat. Neurosci. 2013, 16, 1211–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traiffort, E.; Kassoussi, A.; Zahaf, A.; Laouarem, Y. Astrocytes and Microglia as Major Players of Myelin Production in Normal and Pathological Conditions. Front. Cell. Neurosci. 2020, 14, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- HDomingues, H.S.; Portugal, C.C.; Socodato, R.; Relvas, J.B. Oligodendrocyte, Astrocyte, and Microglia Crosstalk in Myelin Development, Damage, and Repair. Front. Cell Dev. Biol. 2016, 4, 71. [Google Scholar] [CrossRef] [Green Version]
- Martinsen, V.; Kursula, P. Multiple sclerosis and myelin basic protein: Insights into protein disorder and disease. Amino Acids 2021, 54, 99–109. [Google Scholar] [CrossRef]
- Hoehn, H.J.; Kress, Y.; Sohn, A.; Brosnan, C.F.; Bourdon, S.; Shafit-Zagardo, B. Axl−/− mice have delayed recovery and prolonged axonal damage following cuprizone toxicity. Brain Res. 2008, 1240, 1–11. [Google Scholar] [CrossRef]
- Ray, A.K.; Dubois, J.C.; Gruber, R.C.; Guzik, H.M.; Gulinello, M.E.; Perumal, G.; Raine, C.; Kozakiewicz, L.; Williamson, J.; Shafit-Zagardo, B. Loss of Gas6 and Axl signaling results in extensive axonal damage, motor deficits, prolonged neuroinflammation, and less remyelination following cuprizone exposure. Glia 2017, 65, 2051–2069. [Google Scholar] [CrossRef]
- Saher, G.; Brügger, B.; Lappe-Siefke, C.; Möbius, W.; Tozawa, R.-I.; Wehr, M.C.; Wieland, F.; Ishibashi, S.; Nave, K.-A. High cholesterol level is essential for myelin membrane growth. Nat. Neurosci. 2005, 8, 468–475. [Google Scholar] [CrossRef]
- Shafit-Zagardo, B.; Gruber, R.C.; DuBois, J.C. The role of TAM family receptors and ligands in the nervous system: From development to pathobiology. Pharmacol. Ther. 2018, 188, 97–117. [Google Scholar] [CrossRef]
- Coughlin, S.; Noviski, M.; Mueller, J.L.; Chuwonpad, A.; Raschke, W.C.; Weiss, A.; Zikherman, J. An extracatalytic function of CD45 in B cells is mediated by CD22. Proc. Natl. Acad. Sci. USA 2015, 112, E6515–E6524. [Google Scholar] [CrossRef] [Green Version]
- Vodrazka, P.; Korostylev, A.; Hirschberg, A.; Swiercz, J.M.; Worzfeld, T.; Deng, S.; Fazzari, P.; Tamagnone, L.; Offermanns, S.; Kuner, R. The semaphorin 4D-plexin-B signaling complex regulates dendritic and axonal complexity in developing neurons via diverse pathways. Eur. J. Neurosci. 2009, 30, 1193–1208. [Google Scholar] [CrossRef] [PubMed]
- Ebnet, K. Junctional Adhesion Molecules (JAMs): Cell Adhesion Receptors With Pleiotropic Functions in Cell Physiology and Development. Physiol. Rev. 2017, 97, 1529–1554. [Google Scholar] [CrossRef] [PubMed]
- Scheld, M.; Fragoulis, A.; Nyamoya, S.; Zendedel, A.; Denecke, B.; Krauspe, B.; Teske, N.; Kipp, M.; Beyer, C.; Clarner, T. Mitochondrial Impairment in Oligodendroglial Cells Induces Cytokine Expression and Signaling. J. Mol. Neurosci. 2018, 67, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Amstad, A.; Coray, M.; Frick, C.; Barro, C.; Oechtering, J.; Amann, M.; Wischhusen, J.; Kappos, L.; Naegelin, Y.; Kuhle, J.; et al. Growth differentiation factor 15 is increased in stable MS. Neurol.-Neuroimmunol. Neuroinflamm. 2020, 7, e675. [Google Scholar] [CrossRef] [Green Version]
- Golakani, M.H.; Mohammad, M.G.; Li, H.; Gamble, J.; Breit, S.N.; Ruitenberg, M.J.; Brown, D.A. MIC-1/GDF15 Overexpression Is Associated with Increased Functional Recovery in Traumatic Spinal Cord Injury. J. Neurotrauma 2019, 36, 3410–3421. [Google Scholar] [CrossRef]
- Araki, T.; Milbrandt, J. Ninjurin2, a Novel Homophilic Adhesion Molecule, Is Expressed in Mature Sensory and Enteric Neurons and Promotes Neurite Outgrowth. J. Neurosci. 2000, 20, 187–195. [Google Scholar] [CrossRef] [Green Version]
- Allen, M.; Wang, X.; Burgess, J.D.; Watzlawik, J.; Serie, D.J.; Younkin, C.S.; Nguyen, T.; Malphrus, K.G.; Lincoln, S.; Carrasquillo, M.M.; et al. Conserved brain myelination networks are altered in Alzheimer’s and other neurodegenerative diseases. Alzheimer’s Dement. 2017, 14, 352–366. [Google Scholar] [CrossRef]
- YShimotsuma, Y.; Tanaka, M.; Izawa, T.; Yamate, J.; Kuwamura, M. Enhanced Expression of Trib3 during the Development of Myelin Breakdown in dmy Myelin Mutant Rats. PLoS ONE 2016, 11, e0168250. [Google Scholar] [CrossRef] [Green Version]
- Abe, H.; Tanaka, T.; Kimura, M.; Mizukami, S.; Saito, F.; Imatanaka, N.; Akahori, Y.; Yoshida, T.; Shibutani, M. Cuprizone decreases intermediate and late-stage progenitor cells in hippocampal neurogenesis of rats in a framework of 28-day oral dose toxicity study. Toxicol. Appl. Pharmacol. 2015, 287, 210–221. [Google Scholar] [CrossRef]
- Bruce, K.D.; Gorkhali, S.; Given, K.; Coates, A.M.; Boyle, K.E.; Macklin, W.B.; Eckel, R.H. Lipoprotein Lipase Is a Feature of Alternatively-Activated Microglia and May Facilitate Lipid Uptake in the CNS During Demyelination. Front. Mol. Neurosci. 2018, 11, 57. [Google Scholar] [CrossRef] [Green Version]
- Gaultier, A.; Wu, X.; Le Moan, N.; Takimoto, S.; Mukandala, G.; Akassoglou, K.; Campana, W.M.; Gonias, S.L. Low-density lipoprotein receptor-related protein 1 is an essential receptor for myelin phagocytosis. J. Cell Sci. 2009, 122, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Hoyos, H.; Rinaldi, M.; Mendez-Huergo, S.; Marder, M.; Rabinovich, G.; Pasquini, J.; Pasquini, L. Galectin-3 controls the response of microglial cells to limit cuprizone-induced demyelination. Neurobiol. Dis. 2014, 62, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Deczkowska, A.; Keren-Shaul, H.; Weiner, A.; Colonna, M.; Schwartz, M.; Amit, I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 2018, 173, 1073–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, L.; Pasquini, L.A. Galectin-3 Exerts a Pro-differentiating and Pro-myelinating Effect Within a Temporal Window Spanning Precursors and Pre-oligodendrocytes: Insights into the Mechanisms of Action. Mol. Neurobiol. 2020, 57, 976–987. [Google Scholar] [CrossRef]
- Mason, J.L.; Angelastro, J.M.; Ignatova, T.N.; Kukekov, V.G.; Lin, G.; Greene, L.A.; Goldman, J.E. ATF5 regulates the proliferation and differentiation of oligodendrocytes. Mol. Cell. Neurosci. 2005, 29, 372–380. [Google Scholar] [CrossRef]
- Shen, K.; Reichelt, M.; Kyauk, R.V.; Ngu, H.; Shen, Y.-A.A.; Foreman, O.; Modrusan, Z.; Friedman, B.A.; Sheng, M.; Yuen, T.J. Multiple sclerosis risk gene Mertk is required for microglial activation and subsequent remyelination. Cell Rep. 2021, 34, 108835. [Google Scholar] [CrossRef]
- Raddatz, B.B.; Sun, W.; Brogden, G.; Sun, Y.; Kammeyer, P.; Kalkuhl, A.; Colbatzky, F.; Deschl, U.; Naim, H.Y.; Baumgärtner, W.; et al. Central Nervous System Demyelination and Remyelination is Independent from Systemic Cholesterol Level in Theiler’s Murine Encephalomyelitis. Brain Pathol. 2015, 26, 102–119. [Google Scholar] [CrossRef]
- Li, Y.; Xu, X.; Zhang, D.; Cheng, W.; Zhang, Y.; Yu, B.; Zhang, Y. Genetic variation in the leukotriene pathway is associated with myocardial infarction in the Chinese population. Lipids Health Dis. 2019, 18, 25. [Google Scholar] [CrossRef] [Green Version]
- Ellison, J.A.; Barone, F.C.; Feuerstein, G.Z. Matrix Remodeling after Stroke: De Novo Expression of Matrix Proteins and Integrin Receptors. Ann. N. Y. Acad. Sci. 1999, 890, 204–222. [Google Scholar] [CrossRef]
- Wang, X.; Louden, C.; Yue, T.-L.; Ellison, J.A.; Barone, F.C.; Solleveld, H.A.; Feuerstein, G.Z. Delayed expression of osteopontin after focal stroke in the rat. J. Neurosci. 1998, 18, 2075–2083. [Google Scholar] [CrossRef] [Green Version]
- Chabas, D.; Baranzini, S.E.; Mitchell, D.; Bernard, C.C.A.; Rittling, S.R.; Denhardt, D.T.; Sobel, R.A.; Lock, C.; Karpuj, M.; Pedotti, R.; et al. The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science 2001, 294, 1731–1735. [Google Scholar] [CrossRef] [PubMed]
- Selvaraju, R.; Bernasconi, L.; Losberger, C.; Graber, P.; Kadi, L.; Avellana-Adalid, V.; Picard-Riera, N.; Van Evercooren, A.B.; Cirillo, R.; Kosco-Vilbois, M.; et al. Osteopontin is upregulated during in vivo demyelination and remyelination and enhances myelin formation in vitro. Mol. Cell. Neurosci. 2004, 25, 707–721. [Google Scholar] [CrossRef]
- Minor, K.; Phillips, J.; Seeds, N.W. Tissue plasminogen activator promotes axonal outgrowth on CNS myelin after conditioned injury. J. Neurochem. 2009, 109, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Mazaheri, N.; Peymani, M.; Galehdari, H.; Ghaedi, K.; Ghoochani, A.; Kiani-Esfahani, A.; Nasr-Esfahani, M.H. Ameliorating Effect of Osteopontin on H2O2-Induced Apoptosis of Human Oligodendrocyte Progenitor Cells. Cell. Mol. Neurobiol. 2018, 38, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.O.; Mulinyawe, S.B.; Collins, H.Y.; Ibrahim, A.; Li, Q.; Simon, D.J.; Tessier-Lavigne, M.; Barres, B.A. Spatiotemporal Control of CNS Myelination by Oligodendrocyte Programmed Cell Death through the TFEB-PUMA Axis. Cell 2018, 175, 1811–1826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Smaele, E.; Zazzeroni, F.; Papa, S.; Nguyen, D.U.; Jin, R.; Jones, J.; Cong, R.; Franzoso, G. Induction of gadd45beta by NF-kappaB downregulates pro-apoptotic JNK signalling. Nature 2001, 414, 308–313. [Google Scholar] [CrossRef]
- Zhang, J.-X.; Feng, Y.-F.; Qi, Q.; Shen, L.; Wang, R.; Zhou, J.-S.; Lü, H.-Z.; Hu, J.-G. JNK is necessary for oligodendrocyte precursor cell proliferation induced by the conditioned medium from B104 neuroblastoma cells. J. Mol. Neurosci. 2014, 52, 269–276. [Google Scholar] [CrossRef]
- Solga, A.C.; Pong, W.W.; Walker, J.; Wylie, T.; Magrini, V.; Apicelli, A.J.; Griffith, M.; Griffith, O.L.; Kohsaka, S.; Wu, G.F.; et al. RNA-sequencing reveals oligodendrocyte and neuronal transcripts in microglia relevant to central nervous system disease. Glia 2015, 63, 531–548. [Google Scholar] [CrossRef] [Green Version]
- Caminero, A.; Comabella, M.; Montalban, X. Role of tumour necrosis factor (TNF)-α and TNFRSF1A R92Q mutation in the pathogenesis of TNF receptor-associated periodic syndrome and multiple sclerosis. Clin. Exp. Immunol. 2011, 166, 338–345. [Google Scholar] [CrossRef]
- Kassiotis, G.; Pasparakis, M.; Kollias, G.; Probert, L. TNF accelerates the onset but does not alter the incidence and severity of myelin basic protein-induced experimental autoimmune encephalomyelitis. Eur. J. Immunol. 1999, 29, 774–780. [Google Scholar] [CrossRef]
- Fortin, J.; Tian, R.; Zarrabi, I.; Hill, G.; Williams, E.; Sanchez-Duffhues, G.; Thorikay, M.; Ramachandran, P.; Siddaway, R.; Wong, J.F.; et al. Mutant ACVR1 Arrests Glial Cell Differentiation to Drive Tumorigenesis in Pediatric Gliomas. Cancer Cell 2020, 37, 308–323.e12. [Google Scholar] [CrossRef] [PubMed]
- Quan, L.; Uyeda, A.; Muramatsu, R. Central nervous system regeneration: The roles of glial cells in the potential molecular mechanism underlying remyelination. Inflamm. Regen. 2022, 42, 7. [Google Scholar] [CrossRef] [PubMed]
- Auderset, L.; Pitman, K.A.; Cullen, C.L.; Pepper, R.E.; Taylor, B.V.; Foa, L.; Young, K.M. Low-Density Lipoprotein Receptor-Related Protein 1 (LRP1) Is a Negative Regulator of Oligodendrocyte Progenitor Cell Differentiation in the Adult Mouse Brain. Front. Cell Dev. Biol. 2020, 8, 1067. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, R.W.; Michalski, J.-P.; Kothary, R. Integrin Signaling in Oligodendrocytes and Its Importance in CNS Myelination. J. Signal Transduct. 2011, 2011, 354091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arrate, M.P.; Rodriguez, J.M.; Tran, T.M.; Brock, T.A.; Cunningham, S.A. Cloning of human junctional adhesion molecule 3 (JAM3) and its identification as the JAM2 counter-receptor. J. Biol. Chem. 2001, 276, 45826–45832. [Google Scholar] [CrossRef] [Green Version]
- Liang, T.W.; Chiu, H.H.; Gurney, A.; Sidle, A.; Tumas, D.B.; Schow, P.; Foster, J.; Klassen, T.; Dennis, K.; DeMarco, R.A.; et al. Vascular endothelial-junctional adhesion molecule (VE-JAM)/JAM 2 interacts with T, NK, and dendritic cells through JAM 3. J. Immunol. 2002, 168, 1618–1626. [Google Scholar] [CrossRef] [Green Version]
- Ferent, J.; Zimmer, C.; Durbec, P.; Ruat, M.; Traiffort, E. Sonic Hedgehog Signaling Is a Positive Oligodendrocyte Regulator during Demyelination. J. Neurosci. 2013, 33, 1759–1772. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Zhang, X.; Xu, P.; Zhao, N.; Zhao, Y.; Li, Y.; Hong, Y.; Peng, M.; Yuan, K.; Wan, T.; et al. Aberrant oligodendroglial LDL receptor orchestrates demyelination in chronic cerebral ischemia. J. Clin. Investig. 2021, 131, e128114. [Google Scholar] [CrossRef]
- Pietiäinen, V.; Vassilev, B.; Blom, T.; Wang, W.; Nelson, J.; Bittman, R.; Bäck, N.; Zelcer, N.; Ikonen, E. NDRG1 functions in LDL receptor trafficking by regulating endosomal recycling and degradation. J. Cell Sci. 2013, 126, 3961–3971. [Google Scholar] [CrossRef] [Green Version]
- Furusho, M.; Dupree, J.L.; Nave, K.-A.; Bansal, R. Fibroblast Growth Factor Receptor Signaling in Oligodendrocytes Regulates Myelin Sheath Thickness. J. Neurosci. 2012, 32, 6631–6641. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Zhang, X.; Zhang, L.; Xu, D.; Cheng, N.; Tang, Y.; Peng, Y. Astrocytes increase exosomal secretion of oligodendrocyte precursor cells to promote their proliferation via integrin β4-mediated cell adhesion. Biochem. Biophys. Res. Commun. 2020, 526, 341–348. [Google Scholar] [CrossRef] [PubMed]
- van der Zee, C.E.E.M.; Kreft, M.; Beckers, G.; Kuipers, A.; Sonnenberg, A. Conditional Deletion of the Itgb4 Integrin Gene in Schwann Cells Leads to Delayed Peripheral Nerve Regeneration. J. Neurosci. 2008, 28, 11292–11303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darling, T.K.; Lamb, T.J. Emerging Roles for Eph Receptors and Ephrin Ligands in Immunity. Front. Immunol. 2019, 10, 1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linneberg, C.; Harboe, M.; Laursen, L.S. Axo-Glia Interaction Preceding CNS Myelination Is Regulated by Bidirectional Eph-Ephrin Signaling. ASN Neuro 2015, 7, 1759091415602859. [Google Scholar] [CrossRef] [PubMed]
- Diaz, B.G.; Riquelme, R.; Varela-Nieto, I.; Jiménez, A.J.; De Diego, I.; Gómez-Conde, A.I.; Matas-Rico, E.; Aguirre, J.; Chun, J.; Pedraza, C.; et al. Loss of lysophosphatidic acid receptor LPA1 alters oligodendrocyte differentiation and myelination in the mouse cerebral cortex. Brain Struct. Funct. 2015, 220, 3701–3720. [Google Scholar] [CrossRef]
- Akkermann, R.; Aprico, A.; Perera, A.A.; Bujalka, H.; Cole, A.E.; Xiao, J.; Field, J.; Kilpatrick, T.J.; Binder, M.D. The TAM receptor Tyro3 regulates myelination in the central nervous system. Glia 2017, 65, 581–591. [Google Scholar] [CrossRef]
- Martin, N.A.; Nawrocki, A.; Molnar, V.; Elkjaer, M.L.; Thygesen, E.K.; Palkovits, M.; Acs, P.; Sejbaek, T.; Nielsen, H.H.; Hegedus, Z.; et al. Orthologous proteins of experimental de- and remyelination are differentially regulated in the CSF proteome of multiple sclerosis subtypes. PLoS ONE 2018, 13, e0202530. [Google Scholar] [CrossRef] [Green Version]
- Poliani, P.L.; Wang, Y.; Fontana, E.; Robinette, M.L.; Yamanishi, Y.; Gilfillan, S.; Colonna, M. TREM2 sustains microglial expansion during aging and response to demyelination. J. Clin. Investig. 2015, 125, 2161–2170. [Google Scholar] [CrossRef] [Green Version]
- Moyon, S.; Dubessy, A.L.; Aigrot, M.S.; Trotter, M.; Huang, J.K.; Dauphinot, L.; Potier, M.C.; Kerninon, C.; Parsadaniantz, S.M.; Franklin, R.J.; et al. Demyelination causes adult CNS progenitors to revert to an immature state and express immune cues that support their migration. J. Neurosci. 2015, 35, 4–20. [Google Scholar] [CrossRef] [Green Version]
- Han, M.H.; Lundgren, D.H.; Jaiswal, S.; Chao, M.; Graham, K.L.; Garris, C.S.; Axtell, R.C.; Ho, P.P.; Lock, C.B.; Woodard, J.I. Janus-like opposing roles of CD47 in autoimmune brain inflammation in humans and mice. J. Exp. Med. 2012, 209, 1325–1334. [Google Scholar] [CrossRef]
- Kipp, M.; Nyamoya, S.; Hochstrasser, T.; Amor, S. Multiple sclerosis animal models: A clinical and histopathological perspective. Brain Pathol. 2017, 27, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Lieury, A.; Chanal, M.; Androdias, G.; Reynolds, R.; Cavagna, S.; Giraudon, P.; Confavreux, C.; Nataf, S. Tissue remodeling in periplaque regions of multiple sclerosis spinal cord lesions. Glia 2014, 62, 1645–1658. [Google Scholar] [CrossRef] [PubMed]
- Steen, C.B.; Liu, C.L.; Alizadeh, A.A.; Newman, A.M. Profiling Cell Type Abundance and Expression in Bulk Tissues with CIBERSORTx. In Stem Cell Transcriptional Networks, Methods and Protocols; Kidder, B.L., Ed.; Humana: New York, NY, USA, 2020; Volume 2117, pp. 135–157. [Google Scholar]
- Donovan, M.K.R.; D’Antonio-Chronowska, A.; D’Antonio, M.; Frazer, K.A. Cellular deconvolution of GTEz tissues powers Discovery of disease and cell-type associated regulatory variants. Nat. Commun. 2020, 11, 955. [Google Scholar] [CrossRef] [Green Version]
- Browaeys, R.; Saelens, W.; Saeys, Y. NicheNet: Modeling intercellular communication by linking ligands to target genes. Nat. Methods 2020, 17, 159–162. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enrich-Bengoa, J.; Manich, G.; Dégano, I.R.; Perálvarez-Marín, A. Deciphering the Genetic Crosstalk between Microglia and Oligodendrocyte Precursor Cells during Demyelination and Remyelination Using Transcriptomic Data. Int. J. Mol. Sci. 2022, 23, 14868. https://doi.org/10.3390/ijms232314868
Enrich-Bengoa J, Manich G, Dégano IR, Perálvarez-Marín A. Deciphering the Genetic Crosstalk between Microglia and Oligodendrocyte Precursor Cells during Demyelination and Remyelination Using Transcriptomic Data. International Journal of Molecular Sciences. 2022; 23(23):14868. https://doi.org/10.3390/ijms232314868
Chicago/Turabian StyleEnrich-Bengoa, Jennifer, Gemma Manich, Irene R. Dégano, and Alex Perálvarez-Marín. 2022. "Deciphering the Genetic Crosstalk between Microglia and Oligodendrocyte Precursor Cells during Demyelination and Remyelination Using Transcriptomic Data" International Journal of Molecular Sciences 23, no. 23: 14868. https://doi.org/10.3390/ijms232314868