Probing the Efficiency of 13-Pyridylalkyl Berberine Derivatives to Human Telomeric G-Quadruplexes Binding: Spectroscopic, Solid State and In Silico Analysis
Abstract
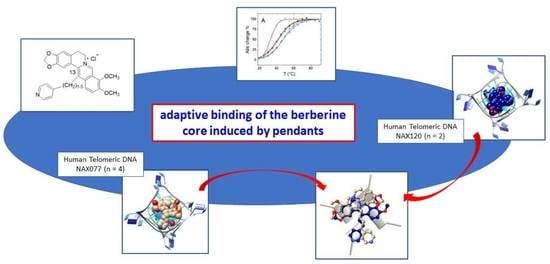
:1. Introduction
2. Results and Discussion
2.1. Solution Studies
2.2. Solid State Study
2.3. Hirshfeld Surface’s Analysis
2.4. Molecular Modeling Study
3. Materials and Methods
3.1. Materials
3.2. Solution Studies
3.3. X-ray Diffraction Analysis—Crystallization
3.4. X-ray Diffraction Analysis—Data Collection and Structure Solution and Refinement
3.5. In Silico Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Phan, A.T.; Kuryavyi, V.; Patel, D.J. DNA architecture: From G to Z. Curr. Opin. Struct. Biol. 2006, 16, 288–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lightfoot, H.L.; Hagen, T.; Tatum, N.J.; Hall, J. The diverse structural landscape of quadruplexes. FEBS Lett. 2019, 593, 2083–2102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, E.; Hardin, C.C.; Walk, S.K.; Tinoco, I., Jr.; Blackburn, E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell 1987, 51, 899–908. [Google Scholar] [CrossRef]
- Sundquist, W.I.; Klug, A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 1989, 342, 825–829. [Google Scholar] [CrossRef]
- Kwok, C.K.; Merrick, C.J. G-quadruplexes: Prediction, characterization, and biological application. Trends Biotechnol. 2017, 35, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Lejault, P.; Chevrier, S.; Boidot, R.; Robertson, A.G.; Wong, J.M.Y.; Monchaud, D. Transcriptome-wide identification of transient RNA Gquadruplexes in human cells. Nat. Commun. 2018, 9, 4730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thandapani, P.; Song, J.; Gandin, V.; Cai, Y.; Rouleau, S.G.; Garant, J.M.; Boisvert, F.M.; Yu, Z.; Perreault, J.P.; Topisirovic, I.; et al. Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. eLife 2015, 4, 06234. [Google Scholar] [CrossRef] [PubMed]
- Murat, P.; Zhong, J.; Lekieffre, L.; Cowieson, N.P.; Clancy, J.L.; Preiss, T.; Balasubramanian, S.; Khanna, R.; Tellam, J. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014, 10, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Agarwala, P.; Pandey, S.; Mapa, K.; Maiti, S. The G-quadruplex augments translation in the 5′ untranslated region of transforming growth factor beta2. Biochemistry 2013, 52, 1528–1538. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armas, P.; David, A.; Calcaterra, N.B. Transcriptional control by G-quadruplexes: In vivo roles and perspectives for specific intervention. Transcription 2017, 8, 21–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valton, A.L.; Prioleau, M.N. G-quadruplexes in DNA replication: A problem or a necessity? Trends Genet. 2016, 32, 697–706. [Google Scholar] [CrossRef]
- Mao, S.Q.; Ghanbarian, A.T.; Spiegel, J.; Martinez Cuesta, S.; Beraldi, D.; Di Antonio, M.; Marsico, G.; Hansel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. DNA G-quadruplex structures mould the DNA methylome. Nat. Struct. Mol. Biol. 2018, 25, 951–957. [Google Scholar] [CrossRef]
- Hansel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, J.; Wild, A.T.; Wu, W.H.; Shah, R.; Danussi, C.; Riggins, G.J.; Kannan, K.; Sulman, E.P.; Chan, T.A.; et al. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat. Commun. 2019, 10, 943. [Google Scholar] [CrossRef] [Green Version]
- Cammas, A.; Millevoi, S. RNA Gquadruplexes: Emerging mechanisms in disease. Nucleic Acids Res. 2016, 45, 1584–1595. [Google Scholar] [CrossRef] [Green Version]
- Maizels, N. G4-associated human diseases. EMBO Rep. 2015, 16, 910–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, R.; Miller, K.M.; Forment, J.V.; Bradshaw, C.R.; Nikan, M.; Britton, S.; Oelschlaegel, T.; Xhemalce, B.; Balasubramanian, S.; Jackson, S.P. Small-molecule–induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8, 301–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagatsugi, F.; Onizuka, K. Functional G-Quadruplex Binding Molecules. Chem. Lett. 2020, 49, 771–780. [Google Scholar] [CrossRef]
- Asamitsu, S.; Bando, T.; Sugiyama, H. Ligand Design to Acquire Specificity to Intended G-Quadruplex Structures. Chem.-Eur. J. 2019, 25, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.R.; Cadoni, E.; Ressurreição, A.S.; Moreira, R.; Paulo, A. Design of Modular G-quadruplex Ligands. ChemMedChem 2018, 13, 869–893. [Google Scholar] [CrossRef]
- Monsen, R.C.; Trent, J.O. G-quadruplex Virtual Drug Screening: A Review. Biochimie 2018, 152, 134–148. [Google Scholar] [CrossRef]
- Macii, F.; Perez-Arnaiz, C.; Arrico, L.; Busto, N.; Garcia, B.; Biver, T. Alcian blue pyridine variant interaction with DNA and RNA polynucleotides and G-quadruplexes: Changes in the binding features for different biosubstrates. J. Inorg. Biochem. 2020, 212, 111199. [Google Scholar] [CrossRef] [PubMed]
- Macii, F.; Cupellini, L.; Stifano, M.; Santolaya, J.; Pérez-Arnaiz, C.; Pucci, A.; Barone, G.; García, B.; Busto, N.; Biver, T. Combined spectroscopic and theoretical analysis of the binding of a water-soluble perylene diimide to DNA/RNA polynucleotides and G-quadruplexes. Spectrochim. Acta A 2021, 260, 119914. [Google Scholar] [CrossRef] [PubMed]
- Biver, T. Discriminating between Parallel, Anti-Parallel and Hybrid G-Quadruplexes: Mechanistic Details on Their Binding to Small Molecules. Molecules 2022, 27, 4165. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [Green Version]
- Todd, A.K.; Johnston, M.; Neidle, S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wang, F.; Song, Y.; Liu, H. The status of and trends in the pharmacology of berberine: A bibliometric review [1985–2018]. Chin. Med. 2020, 15, 7. [Google Scholar] [CrossRef] [Green Version]
- Chu, M.; Chen, X.; Wang, J.; Guo, L.; Wang, Q.; Gao, Z.; Kang, J.; Zhang, M.; Feng, J.; Guo, Q.; et al. Polypharmacology of Berberine Based on Multi-Target Binding Motifs. Front. Pharmacol. 2018, 9, 801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, F.; Xie, T.; Huang, X.; Zhao, X. Berberine inhibits angiogenesis in glioblastoma xenografts by targeting the VEGFR2/ERK pathway. Pharm. Biol. 2018, 56, 665–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhang, S. Berberine suppresses growth and metastasis of endometrial cancer cells via miR-101/COX-2 BIOMED. Biomed. Pharmacother. 2018, 103, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.-C.; Yu, C.-C.; Hsu, L.-S.; Chen, K.-S.; Su, M.-Y.; Chen, P.-N. Berberine Reverses Epithelial-to-Mesenchymal Transition and Inhibits Metastasis and Tumor-Induced Angiogenesis in Human Cervical Cancer Cells. Mol. Pharmacol. 2014, 86, 609–623. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.-H.; Lo, H.-L.; Tang, W.-C.; Hsiao, H.H.-Y.; Yang, P. A gene expression signature-based approach reveals the mechanisms of action of the Chinese herbal medicine berberine. Sci. Rep. 2014, 4, 6394. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Ou, T.-M.; Hou, J.-Q.; Lu, Y.-J.; Tan, J.-H.; Gu, L.-Q.; Huang, Z.-S. 9-N-Substituted Berberine Derivatives: Stabilization of G-Quadruplex DNA and down-Regulation of Oncogene c-Myc. Bioorg. Med. Chem. 2008, 16, 7582–7591. [Google Scholar] [CrossRef]
- Arora, A.; Balasubramanian, C.; Kumar, N.; Agrawal, S.; Ojha, R.P.; Maiti, S. Binding of Berberine to Human Telomeric Quadruplex—Spectroscopic, Calorimetric and Molecular Modeling Studies. FEBS J. 2008, 275, 3971–3983. [Google Scholar] [CrossRef] [PubMed]
- Ferraroni, M.; Bazzicalupi, C.; Papi, F.; Fiorillo, G.; Guaman-Ortiz, L.M.; Nocentini, A.; Scovassi, A.I.; Lombardi, P.; Gratteri, P. Solution and Solid-State Analysis of Binding of 13-Substituted Berberine Analogues to Human Telomeric G-quadruplexes. Chem.-Asian J. 2016, 11, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Bazzicalupi, C.; Ferraroni, M.; Bilia, A.R.; Scheggi, F.; Gratteri, P. The crystal structure of human telomeric DNA complexed with berberine: An interesting case of stacked ligand to G-tetrad ratio higher than 1:1. Nucleic Acids Res. 2013, 41, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, C.; Liu, Y.; Lan, W.; Han, H.; Wang, R.; Huang, S.; Cao, C. Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR. Chem. Commun. 2020, 56, 2099–2102. [Google Scholar] [CrossRef]
- Chatterjee, S.; Mallick, S.; Buzzetti, F.; Fiorillo, G.; Syeda, T.M.; Lombardi, P.; Saha, K.D.; Kumar, G.S. New 13-pyridinealkyl berberine analogues intercalate to DNA and induce apoptosis in HepG2 and MCF-7 cells through ROS mediated p53 dependent pathway: Biophysical, biochemical and molecular modeling studies. RSC Adv. 2015, 5, 90632–90644. [Google Scholar] [CrossRef]
- Papi, F.; Bazzicalupi, C.; Ferraroni, M.; Ciolli, G.; Lombardi, P.; Khan, A.Y.; Kumar, G.S.; Gratteri, P. Pyridine Derivative of the Natural Alkaloid Berberine as Human Telomeric G4-DNA Binder: A Solution and Solid-State Study. ACS Med. Chem. Lett. 2020, 11, 645–650. [Google Scholar] [CrossRef]
- Available online: http://www.hyperquad.co.uk (accessed on 1 August 2022).
- Papi, F.; Ferraroni, M.; Rigo, R.; Da Ros, S.; Bazzicalupi, C.; Sissi, C.; Gratteri, P. Role of the Benzodioxole Group in the Interactions between the Natural Alkaloids Chelerythrine and Coptisine and the Human Telomeric G-Quadruplex DNA. A Multiapproach Investigation. J. Nat. Prod. 2017, 80, 3128–3135. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Cuenca, F.; Neidle, S. Topology Conservation and Loop Flexibility in Quadruplex–Drug Recognition: Crystal Structures of Inter- and Intramolecular Telomeric DNA Quadruplex–Drug Complexes. J. Mol. Biol. 2008, 381, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Collie, G.W.; Campbell, N.H.; Neidle, S. Loop flexibility in human telomeric quadruplex small-molecule complexes. Nucleic Acids Res. 2015, 43, 4785–4799. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 37, 3814–3816. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Mitchell, A.S.; Spackman, M.A. Molecular surfaces from the promolecule: A comparison with Hartree–Fock ab initio electron density surfaces. J. Comput. Chem. 2000, 21, 933–942. [Google Scholar] [CrossRef]
- Spackman, M.A.; Maslen, E.N. Chemical properties from the promolecule. J. Phys. Chem. 1986, 90, 2020–2027. [Google Scholar] [CrossRef]
- Ortiz de Luzuriaga, I.; Lopez, X.; Gil, A. Learning to Model G-Quadruplexes: Current Methods and Perspectives. Annu. Rev. Biophys. 2021, 50, 209–243. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Dai, J.X.; Punchihewa, C.; Ambrus, A.; Chen, D.; Jones, R.A.; Yang, D.Z. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: A novel adenine triple formation. Nucleic Acids Res. 2007, 35, 2440–2450. [Google Scholar] [CrossRef] [Green Version]
- Iwasa, K.; Kamigauchi, M.; Sugiura, M.; Nanba, H. Antimicrobial activity of some 13-alkyl substituted protoberberinium salts. Planta Med. 1997, 63, 196–198. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Phan, A.-T.; Lacroix, L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998, 435, 74–78. [Google Scholar] [CrossRef] [Green Version]
- Campbell, N.H.; Parkinson, G.N. Crystallographic studies of quadruplex nucleic acids. Methods 2007, 43, 252–263. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D 2010, 66, 125–132. [Google Scholar] [CrossRef] [Green Version]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. J. Acta Crystallogr. D 1997, 53, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Kee-gan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 2011, 67, 235–242. [Google Scholar] [CrossRef] [Green Version]
- GRADE 1.2.12 (4 October 2016). Available online: http://grade.globalphasing.org (accessed on 16 July 2021).
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K.D. Features and development of Coot. Acta Crystallogr. D 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; van de Streek, J.; Wood, P.A. Mercury CSD 2.0—New features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Wirmer-Bartoschek, J.; Bendel, L.E.; Jonker, H.R.A.; Grun, J.T.; Papi, F.; Bazzicalupi, C.; Messori, L.; Gratteri, P.; Schwalbe, H. Solution NMR Structure of a Ligand/Hybrid-2-G-Quadruplex Complex Reveals Rearrangements that Affect Ligand Binding. Angew. Chem. Int. Edit. 2017, 56, 7102–7106. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger Suite Release 2021-3, Schrodinger, LLC, New York, NY, 2019 (Maestro v.12.9; Epik, v.5.7; Macromodel v.13.3; Glide, v.9.2; Desmond, v.6.7; Prime, v.5.5; Impact, v.9.2; Jaguar, v.11.3). Available online: http://www.schrodinger.com (accessed on 12 April 2022).
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, T.-M.; Lu, Y.-J.; Tan, J.-H.; Huang, Z.-S.; Wong, K.-Y.; Gu, L.-Q. G-quadruplexes: Targets in anticancer drug design. ChemMedChem 2008, 3, 690–713. [Google Scholar] [CrossRef]
- Lopes-Nunes, J.; Carvalho, J.; Figueiredo, J.; Ramos, C.I.V.; Lourenço, L.M.O.; Tomé, J.P.C.; Neves, M.G.P.M.S.; Mergny, J.L.; Queiroz, J.A.; Salgado, G.F.; et al. Phthalocyanines for G-Quadruplex Aptamers Binding. Bioorg. Chem. 2020, 100, 103920. [Google Scholar] [CrossRef]
- Li, Z.-Q.; Liao, T.-C.; Dong, C.; Yang, J.-W.; Chen, X.-J.; Liu, L.; Luo, Y.; Liang, Y.-Y.; Chen, W.-H.; Zhou, C.-Q. Specifically targeting mixed-type dimeric G-quadruplexes using berberine dimers. Org. Biomol. Chem. 2017, 15, 10221–10229. [Google Scholar] [CrossRef]








| Tel12 | Tel23 | ||||||
|---|---|---|---|---|---|---|---|
| λmax Free (nm) | λmax Bound (nm) | LogK | ΔTm (°C) | λmax Bound (nm) | LogK | ΔTm (°C) | |
| 1 | 341, 414 | 347, 400, 420 | 6.0 ± 0.2 | 14.4 ± 1.5 | 343 (413 + 445) * | 4.4 ± 0.2 | 4.1 ± 0.4 |
| 2 | 341, 414 | 348, 426 | 6.1 ± 0.2 | 9.7 ± 1.8 | 344, 426 | 4.0 ± 0.3 | 4.1 ± 0.8 |
| 3 | 339, 413 | 349, 425 | 6.6 ± 0.2 | 16.7 ± 1.3 | 350, 427 | 5.8 ± 0.2 | 2.3 ± 0.6 |
| 4 | 338, 413 | 349, 428 | 7.0 ± 0.2 | 8.7 ± 1.5 | 341, 422 | 5.2 ± 0.2 | 5.9 ± 1.0 |
| 5 | 338, 414 | 348, 428 | 7.2 ± 0.2 | 15.0 ± 1.4 | 341, 422 | 5.6 ± 0.2 | 5.1 ± 1.1 |
| Ligand/Interacting Group | Type of Interaction/Involved Residue | |||||
|---|---|---|---|---|---|---|
| π-Stacking (≈3.4 Å) | Medium Long π-Stacking (≈3.6–3.7 Å) | Long π-Stacking (≈3.8–4.0 Å) | NH····O | CH····π | ||
| 2 | Quinoline | G4 | A3 | |||
| Benzyl | ||||||
| Pyridine | ||||||
| Benzodioxole | A15 (2.3 Å) | |||||
| 3 | Quinoline | G22; A15 | ||||
| Benzyl | G4; A3 | T14 | ||||
| Pyridine | ||||||
| Benzodioxole | ||||||
| 4 | Quinoline | T13 | G16 | |||
| Benzyl | G22 | A15 | ||||
| Pyridine | A3 | |||||
| Benzodioxole | A3 (2.2 Å) | |||||
| 5 | Quinoline | A3 | G22 | |||
| Benzyl | A15 | G16 | ||||
| Pyridine | T13 | |||||
| Benzodioxole | T13 (1.9 Å) | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bazzicalupi, C.; Bonardi, A.; Biver, T.; Ferraroni, M.; Papi, F.; Savastano, M.; Lombardi, P.; Gratteri, P. Probing the Efficiency of 13-Pyridylalkyl Berberine Derivatives to Human Telomeric G-Quadruplexes Binding: Spectroscopic, Solid State and In Silico Analysis. Int. J. Mol. Sci. 2022, 23, 14061. https://doi.org/10.3390/ijms232214061
Bazzicalupi C, Bonardi A, Biver T, Ferraroni M, Papi F, Savastano M, Lombardi P, Gratteri P. Probing the Efficiency of 13-Pyridylalkyl Berberine Derivatives to Human Telomeric G-Quadruplexes Binding: Spectroscopic, Solid State and In Silico Analysis. International Journal of Molecular Sciences. 2022; 23(22):14061. https://doi.org/10.3390/ijms232214061
Chicago/Turabian StyleBazzicalupi, Carla, Alessandro Bonardi, Tarita Biver, Marta Ferraroni, Francesco Papi, Matteo Savastano, Paolo Lombardi, and Paola Gratteri. 2022. "Probing the Efficiency of 13-Pyridylalkyl Berberine Derivatives to Human Telomeric G-Quadruplexes Binding: Spectroscopic, Solid State and In Silico Analysis" International Journal of Molecular Sciences 23, no. 22: 14061. https://doi.org/10.3390/ijms232214061