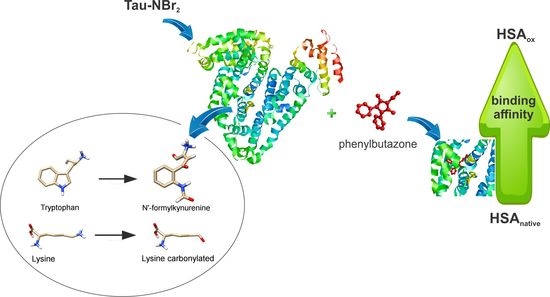
Oxidative Alteration of Trp-214 and Lys-199 in Human Serum Albumin Increases Binding Affinity with Phenylbutazone: A Combined Experimental and Computational Investigation
Abstract
:1. Introduction
2. Results
2.1. Efficacy of Tau-NBr2 as an Oxidant of Free Aromatic Amino Acids
2.2. Oxidation of Amino-Acid Residues in HSA by Tau-NBr2 and Tau-NHCl
2.3. Alteration in the Binding of Phenylbutazone Provoked by Oxidation of HSA
2.4. Computational Alteration of Trp-214 and Lys-199 and Molecular Docking Simulation
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Oxidation of N-acetyl-l-tryptophan and N-acetyl-l-tyrosine
4.3. Oxidation of HSA
4.4. Determination of Sulfhydryl Residues
4.5. Determination of Carbonyl Products
4.6. Determination of Association Constant
4.7. Molecular Docking Simulations
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pisoschi, A.M.; Pop, A. The role of antioxidants in the chemistry of oxidative stress: A review. Eur. J. Med. Chem. 2015, 97, 55–74. [Google Scholar] [CrossRef] [PubMed]
- Araújo, R.F.F.; Martins, D.B.G.; Borba, M.A.C.S.M. Oxidative Stress and Disease. In A Master Regulator of Oxidative Stress-The Transcription Factor Nrf2; Morales-Gonzales, J.A., Morales-Gonzales, A., Madrigal-Santillan, E.O., Eds.; IntechOpen: London, UK, 2016; pp. 185–199. [Google Scholar]
- Poprac, P.; Jomova, K.; Simunkova, M.; Kollar, V.; Rhodes, C.J.; Valko, M. Targeting Free Radicals in Oxidative Stress-Related Human Diseases. Trends Pharmacol. Sci. 2017, 38, 592–607. [Google Scholar] [CrossRef] [PubMed]
- Moloney, J.N.; Cotter, T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Rani, V.; Deep, G.; Singh, R.K.; Palle, K.; Yadav, U.C.S. Oxidative stress and metabolic disorders: Pathogenesis and therapeutic strategies. Life Sci. 2016, 148, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Neves Carvalho, A.; Firuzi, O.; Joao Gama, M.; van Horssen, J.; Saso, L. Oxidative Stress and Antioxidants in Neurological Diseases: Is There Still Hope? Curr. Drug Targets 2017, 18, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Tramutola, A.; Lanzillotta, C.; Perluigi, M.; Butterfield, D.A. Oxidative stress, protein modification and Alzheimer disease. Brain Res. Bull. 2017, 133, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Tan, H.-Y.; Wang, N.; Zhang, Z.-J.; Lao, L.; Wong, C.-W.; Feng, Y. The Role of Oxidative Stress and Antioxidants in Liver Diseases. Int. J. Mol. Sci. 2015, 16, 26087–26124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niedzielska, E.; Smaga, I.; Gawlik, M.; Moniczewski, A.; Stankowicz, P.; Pera, J.; Filip, M. Oxidative Stress in Neurodegenerative Diseases. Mol. Neurobiol. 2016, 53, 4094–4125. [Google Scholar] [CrossRef] [PubMed]
- Roche, M.; Rondeau, P.; Singh, N.R.; Tarnus, E.; Bourdon, E. The antioxidant properties of serum albumin. FEBS Lett. 2008, 582, 1783–1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, S. Role of human serum albumin and oxidative stress in diabetes. J. Appl. Biotechnol. Bioeng. 2017, 3, 1–5. [Google Scholar] [CrossRef]
- Peters, T. All about Albumin: Biochemistry, Genetics, and Medical Applications; Academic Press: Cambridge, MA, USA, 1996; ISBN 9780125521109. [Google Scholar]
- Kouno, Y.; Anraku, M.; Yamasaki, K.; Okayama, Y.; Iohara, D.; Nakamura, H.; Maruyama, T.; Hirayama, F.; Kragh-Hansen, U.; Otagiri, M. N-acetyl-l-methionine is a superior protectant of human serum albumin against post-translational oxidation as compared to N-acetyl-l-tryptophan. Biochem. Biophys. Rep. 2016, 6, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006; ISBN 9780387463124. [Google Scholar]
- Zhivkova, Z. Studies on Drug—Human Serum Albumin Binding: The Current State of the Matter. Curr. Pharm. Des. 2015, 21, 1817–1830. [Google Scholar] [CrossRef] [PubMed]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. The characterization of two specific drug binding sites on human serum albumin. Mol. Pharmacol. 1975, 11, 824–832. [Google Scholar] [PubMed]
- Ghuman, J.; Zunszain, P.A.; Petitpas, I.; Bhattacharya, A.A.; Otagiri, M.; Curry, S. Structural Basis of the Drug-binding Specificity of Human Serum Albumin. J. Mol. Biol. 2005, 353, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Maciążek-Jurczyk, M.; Szkudlarek, A.; Chudzik, M.; Pożycka, J.; Sułkowska, A. Alteration of human serum albumin binding properties induced by modifications: A review. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 188, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Naldi, M.; Baldassarre, M.; Domenicali, M.; Caraceni, P. Structural and functional integrity of human serum albumin: Analytical approaches and clinical relevance in patients with liver cirrhosis. J. Pharm. Biomed. Anal. 2017, 144, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Oettl, K.; Marsche, G. Redox State of Human Serum Albumin in Terms of Cysteine-34 in Health and Disease. Methods Enzymol. 2010, 474, 181–195. [Google Scholar] [PubMed]
- Sitar, M.E.; Aydin, S.; Cakatay, U. Human serum albumin and its relation with oxidative stress. Clin. Lab. 2013, 59, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Arif, Z.; Neelofar, K.; Arfat, M.Y.; Zaman, A.; Tarannum, A.; Parveen, I.; Ahmad, S.; Khan, M.A.; Badar, A.; Islam, S.N. Hyperglycemia induced reactive species trigger structural changes in human serum albumin of type 1 diabetic subjects. Int. J. Biol. Macromol. 2018, 107, 2141–2149. [Google Scholar] [CrossRef] [PubMed]
- Fujii, R.; Ueyama, J.; Aoi, A.; Ichino, N.; Osakabe, K.; Sugimoto, K.; Suzuki, K.; Hamajima, N.; Wakai, K.; Kondo, T. Oxidized human serum albumin as a possible correlation factor for atherosclerosis in a rural Japanese population: The results of the Yakumo Study. Environ. Health Prev. Med. 2018, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Maciążek-Jurczyk, M.; Sułkowska, A. Spectroscopic analysis of the impact of oxidative stress on the structure of human serum albumin (HSA) in terms of its binding properties. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 136, 265–282. [Google Scholar] [CrossRef] [PubMed]
- Maciążek-Jurczyk, M.; Sułkowska, A.; Równicka-Zubik, J. Alteration of methotrexate binding to human serum albumin induced by oxidative stress. Spectroscopic comparative study. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2016, 152, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Oettl, K.; Birner-Gruenberger, R.; Spindelboeck, W.; Stueger, H.P.; Dorn, L.; Stadlbauer, V.; Putz-Bankuti, C.; Krisper, P.; Graziadei, I.; Vogel, W.; et al. Oxidative albumin damage in chronic liver failure: Relation to albumin binding capacity, liver dysfunction and survival. J. Hepatol. 2013, 59, 978–983. [Google Scholar] [CrossRef] [PubMed]
- Russeva, V.N.; Zhivkova, Z.D. Protein binding of some nonsteroidal anti-inflammatory drugs studied by high-performance liquid affinity chromatography. Int. J. Pharm. 1999, 180, 69–74. [Google Scholar] [CrossRef]
- Ximenes, V.F.; da Fonseca, L.M.; de Almeida, A.C. Taurine bromamine: A potent oxidant of tryptophan residues in albumin. Arch. Biochem. Biophys. 2011, 507, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lee, P.; Liang, S.; Zhou, Z.; Wu, X.; Yang, F.; Liang, H. Structural Basis of Non-Steroidal Anti-Inflammatory Drug Diclofenac Binding to Human Serum Albumin. Chem. Biol. Drug Des. 2015, 86, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Ryan, A.J.; Chung, C.-W.; Curry, S. Crystallographic analysis reveals the structural basis of the high-affinity binding of iophenoxic acid to human serum albumin. BMC Struct. Biol. 2011, 11, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, A.J.; Ghuman, J.; Zunszain, P.A.; Chung, C.; Curry, S. Structural basis of binding of fluorescent, site-specific dansylated amino acids to human serum albumin. J. Struct. Biol. 2011, 174, 84–91. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho Bertozo, L.; Morgon, N.; De Souza, A.; Ximenes, V. Taurine Bromamine: Reactivity of an Endogenous and Exogenous Anti-Inflammatory and Antimicrobial Amino Acid Derivative. Biomolecules 2016, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Möller, M.; Denicola, A. Protein tryptophan accessibility studied by fluorescence quenching. Biochem. Mol. Biol. Educ. 2002, 30, 175–178. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.-C.; Wang, H.-M.; Niu, Q.-X.; Ye, D.-Y.; Liang, G.-W. Binding between Saikosaponin C and Human Serum Albumin by Fluorescence Spectroscopy and Molecular Docking. Molecules 2016, 21, 153. [Google Scholar] [CrossRef] [PubMed]
- Reid, L.O.; Roman, E.A.; Thomas, A.H.; Dántola, M.L. Photooxidation of Tryptophan and Tyrosine Residues in Human Serum Albumin Sensitized by Pterin: A Model for Globular Protein Photodamage in Skin. Biochemistry 2016, 55, 4777–4786. [Google Scholar] [CrossRef] [PubMed]
- Ehrenshaft, M.; Silva, S.O.; Perdivara, I.; Bilski, P.; Sik, R.H.; Chignell, C.F.; Tomer, K.B.; Mason, R.P. Immunological detection of N-formylkynurenine in oxidized proteins. Free Radic. Biol. Med. 2009, 46, 1260–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, P.; Koza, S.; Bouvier, E.S.P. Size-Exclusion Chromatography for the Analysis of Protein Biotherapeutics and their Aggregates. J. Liq. Chromatogr. Relat. Technol. 2012, 35, 2923–2950. [Google Scholar] [PubMed]
- Fasman, G.D. Circular Dichroism and the Conformational Analysis of Biomolecules; Springer Science+business Media: New York, NY, USA, 1996. [Google Scholar]
- Zsila, F.; Bikádi, Z.; Simonyi, M. Probing the binding of the flavonoid, quercetin to human serum albumin by circular dichroism, electronic absorption spectroscopy and molecular modelling methods. Biochem. Pharmacol. 2003, 65, 447–456. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Petrônio, M.S.; Ximenes, V.F. Effects of oxidation of lysozyme by hypohalous acids and haloamines on enzymatic activity and aggregation. Biochim. Biophys. Acta 2012, 1824, 1090–1096. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, Y.; Katsuragi, Y.; Izumi, T.; Sakiyama, F. Fluorescence characteristics of kynurenine and N′-formylkynurenine. Their use as reporters of the environment of tryptophan 62 in hen egg-white lysozyme. J. Biochem. 1982, 92, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Grigoryan, H.; Li, H.; Iavarone, A.T.; Williams, E.R.; Rappaport, S.M. Cys34 Adducts of Reactive Oxygen Species in Human Serum Albumin. Chem. Res. Toxicol. 2012, 25, 1633–1642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalle-Donne, I.; Rossi, R.; Giustarini, D.; Milzani, A.; Colombo, R. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta 2003, 329, 23–38. [Google Scholar] [CrossRef]
- Khosravifarsani, M.; Monfared, A.S.; Pouramir, M.; Zabihi, E. Effects of Fenton Reaction on Human Serum Albumin: An In Vitro Study. Electron. Physician 2016, 8, 2970–2976. [Google Scholar] [CrossRef] [PubMed]
- Ghisaidoobe, A.; Chung, S. Intrinsic Tryptophan Fluorescence in the Detection and Analysis of Proteins: A Focus on Förster Resonance Energy Transfer Techniques. Int. J. Mol. Sci. 2014, 15, 22518–22538. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, D.; Bertucci, C. Induced circular dichroism as a tool to investigate the binding of drugs to carrier proteins: Classic approaches and new trends. J. Pharm. Biomed. Anal. 2015, 113, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Ascoli, G.; Bertucci, C.; Salvadori, P. Stereospecific and competitive binding of drugs to human serum albumin: A difference circular dichroism approach. J. Pharm. Sci. 1995, 84, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Maciążek-Jurczyk, M. Phenylbutazone and ketoprofen binding to serum albumin. Fluorescence study. Pharmacol. Rep. 2014, 66, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Lazaridis, T.; Archontis, G.; Karplus, M. Enthalpic Contribution to Protein Stability: Insights from Atom-Based Calculations and Statistical Mechanics. Adv. Protein Chem. 1995, 47, 231–306. [Google Scholar] [PubMed]
- Jones, C.L.; Dickson, T.; Hayes, R.; Thomas, L. Effects of pH and ionic strength on the thermodynamics of human serum albumin-photosensitizer binding. Thermochim. Acta 2012, 545, 112–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Yan, X.-P.; Chen, C.; Xia, Y.-L.; Jiang, Y. Human Serum Albumin−Mercurial Species Interactions. J. Proteome Res. 2007, 6, 2277–2286. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, S.; Kanayama, A.; Miyamoto, Y. Modification of IκBα by taurine bromamine inhibits tumor necrosis factor α-induced NF-κB activation. Inflamm. Res. 2007, 56, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.L.; Bozeman, P.M.; Jefferson, M.M.; King, C.C. Oxidation of bromide by the human leukocyte enzymes myeloperoxidase and eosinophil peroxidase. Formation of bromamines. J. Biol. Chem. 1995, 270, 2906–2913. [Google Scholar] [CrossRef] [PubMed]
- Riener, C.K.; Kada, G.; Gruber, H.J. Quick measurement of protein sulfhydryls with Ellman’s reagent and with 4,4′-dithiodipyridine. Anal. Bioanal. Chem. 2002, 373, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Azizova, O.A.; Aseychev, A.V.; Beckman, E.M.; Moskvina, S.N.; Skotnikova, O.I.; Smolina, N.V.; Gryzunov, Y.A.; Dobretsov, G.E. Studies of oxidant-induced changes in albumin transport function with a fluorescent probe k-35. Effect of hypochlorite. Bull. Exp. Biol. Med. 2012, 152, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Lang, P.T.; Brozell, S.R.; Mukherjee, S.; Pettersen, E.F.; Meng, E.C.; Thomas, V.; Rizzo, R.C.; Case, D.A.; James, T.L.; Kuntz, I.D. DOCK 6: Combining techniques to model RNA-small molecule complexes. RNA 2009, 15, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Asahi, T.; Nakamura, Y.; Kato, Y.; Osawa, T. Specific role of taurine in the 8-brominated-2′-deoxyguanosine formation. Arch. Biochem. Biophys. 2015, 586, 45–50. [Google Scholar] [CrossRef] [PubMed]









© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertozo, L.D.C.; Tavares Neto, E.; Oliveira, L.C.d.; Ximenes, V.F. Oxidative Alteration of Trp-214 and Lys-199 in Human Serum Albumin Increases Binding Affinity with Phenylbutazone: A Combined Experimental and Computational Investigation. Int. J. Mol. Sci. 2018, 19, 2868. https://doi.org/10.3390/ijms19102868
Bertozo LDC, Tavares Neto E, Oliveira LCd, Ximenes VF. Oxidative Alteration of Trp-214 and Lys-199 in Human Serum Albumin Increases Binding Affinity with Phenylbutazone: A Combined Experimental and Computational Investigation. International Journal of Molecular Sciences. 2018; 19(10):2868. https://doi.org/10.3390/ijms19102868
Chicago/Turabian StyleBertozo, Luiza De Carvalho, Ernesto Tavares Neto, Leandro Cristante de Oliveira, and Valdecir Farias Ximenes. 2018. "Oxidative Alteration of Trp-214 and Lys-199 in Human Serum Albumin Increases Binding Affinity with Phenylbutazone: A Combined Experimental and Computational Investigation" International Journal of Molecular Sciences 19, no. 10: 2868. https://doi.org/10.3390/ijms19102868