Koninginins X-Z, Three New Polyketides from Trichoderma koningiopsis SC-5
Abstract
:1. Introduction
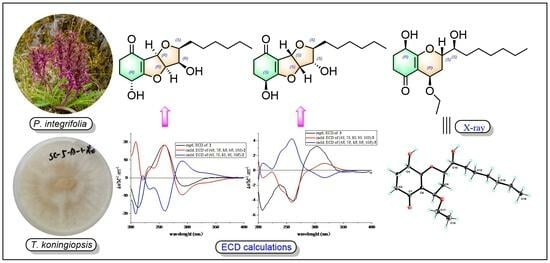
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation, Extraction, and Isolation
3.4. X-ray Crystallographic Analysis
3.5. Quantum Chemistry Calculations
3.6. Biological Activity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.; Pruitt, R.N.; Nürnberger, T.; Wang, Y.C. Evasion of plant immunity by microbial pathogens. Nat. Rev. Microbiol. 2022, 20, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Hartmann, F.E.; Francisco, C.S.; McDonald, B.A.; Croll, D. Mapping the adaptive landscape of a major agricultural pathogen reveals evolutionary constraints across heterogeneous environments. ISME J. 2021, 15, 1402–1419. [Google Scholar] [CrossRef] [PubMed]
- Keswani, C.; Singh, S.P.; García-Estrada, C.; Mezaache-Aichour, S.; Glare, T.R.; Borriss, R.; Rajput, V.D.; Minkina, T.M.; Ortiz, A.; Sansinenea, E. Biosynthesis and beneficial effects of microbial gibberellins on crops for sustainable agriculture. J. Appl. Microbiol. 2022, 132, 1597–1615. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.; Rowlinson, M.; Wang, F.; Bering, L.; Francis, D.; Levy, C.; Micklefield, J. Discovery, characterization and engineering of ligases for amide synthesis. Nature 2021, 593, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.G.; Wang, T.T.; Hua, C.F.; Sun, W.J.; Li, X.Y.; Grunwald, L.; Liu, J.G.; Wu, N.; Shao, X.L.; Yin, Y.M.; et al. A compendium of DNA-binding specificities of transcription factors in Pseudomonas syringae. Nat. Commun. 2020, 11, 4947. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Guo, H.L.; Zhang, K.X.; Zhao, M.Y.; Ruan, J.J.; Chen, J. Trichoderma and its role in biological control of plant fungal and nematode disease. Front. Microbiol. 2023, 14, 1160551. [Google Scholar] [CrossRef]
- Tyśkiewicz, R.; Nowak, A.; Ozimek, E.; Jaroszuk-Ściseł, J. Trichoderma: The current status of its application in agriculture for the biocontrol of fungal phytopathogens and stimulation of plant growth. Int. J. Mol. Sci. 2022, 23, 2329. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.P.; Liang, X.R.; Yin, X.L.; Wang, G.; Ji, N.Y. Absolute configurations of unique harziane diterpenes from Trichoderma species. Org. Lett. 2012, 14, 3815–3817. [Google Scholar] [CrossRef]
- Song, F.H.; Dai, H.Q.; Tong, Y.J.; Ren, B.; Chen, C.X.; Sun, N.; Liu, X.Y.; Bian, J.; Liu, M.; Gao, H.; et al. Trichodermaketones A-D and 7-O-methylkoninginin D from the marine fungus Trichoderma koningii. J. Nat. Prod. 2010, 73, 806–810. [Google Scholar] [CrossRef]
- Sun, Y.; Tian, L.; Huang, J.; Ma, H.Y.; Zheng, Z.; Lv, A.L.; Yasukawa, K.; Pei, Y.H. Trichodermatides A-D, novel polyketides from the marine-derived fungus Trichoderma reesei. Org. Lett. 2008, 10, 393–396. [Google Scholar] [CrossRef]
- Chen, S.C.; Li, H.H.; Chen, Y.C.; Li, S.N.; Xu, J.L.; Guo, H.; Liu, Z.M.; Zhu, S.; Liu, H.X.; Zhang, W.M. Three new diterpenes and two new sesquiterpenoids from the endophytic fungus Trichoderma koningiopsis A729. Bioorg. Chem. 2019, 86, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.S.; Meng, L.H.; Li, X.; Wang, D.J.; Zhou, X.W.; Du, F.Y.; Wang, B.G.; Li, X.M. Polyketides and terpenoids with potent antibacterial activities from the Artemisia argyi-derived fungus Trichoderma koningiopsis QA-3. Chem. Biodivers. 2020, 17, e2000566. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Yang, Y.; Miao, C.P.; Zheng, Y.K.; Chen, J.L.; Chen, Y.W.; Xu, L.H.; Guang, H.L.; Ding, Z.T.; Zhao, L.X. Koningiopisins A-H, polyketides with synergistic antifungal activities from the endophytic fungus Trichoderma koningiopsis. Planta Med. 2016, 82, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wu, G.W.; Liu, D.; Zhuang, W.Y.; Yin, W.B. Trichodermatides E and F from fungus Trichoderma applanatum. J. Asian Nat. Prod. Res. 2019, 21, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Hu, B.Y.; Qian, M.A.; Wang, Z.H.; Zou, J.M.; Sang, X.Y.; Li, L.; Luo, X.D.; Zhao, L.X. Koninginin W, a new polyketide from the endophytic fungus Trichoderma koningiopsis YIM PH30002. Chem. Biodivers. 2021, 18, e2100460. [Google Scholar] [CrossRef]
- Cutler, H.G.; Cutler, S.J.; Ross, S.A.; Sayed, K.E.; Dugan, F.M.; Bartlett, M.G.; Hill, A.A.; Hill, R.A.; Parker, S.R. Koninginin G, a new metabolite from Trichoderma aureoviride. J. Nat. Prod. 1999, 62, 137–139. [Google Scholar] [CrossRef]
- Shi, X.S.; Li, H.L.; Li, X.M.; Wang, D.J.; Li, X.; Meng, L.H.; Zhou, X.W.; Wang, B.G. Highly oxygenated polyketides produced by Trichoderma koningiopsis QA-3, an endophytic fungus obtained from the fresh roots of the medicinal plant Artemisia argyi. Bioorg. Chem. 2020, 94, 103448. [Google Scholar] [CrossRef]
- Hu, M.; Li, Q.L.; Yang, Y.B.; Liu, K.; Miao, C.P.; Zhao, L.X.; Ding, Z.T. Koninginins R-S from the endophytic fungus Trichoderma koningiopsis. Nat. Prod. Res. 2017, 31, 835–839. [Google Scholar] [CrossRef]
- Liu, K.; Yang, Y.B.; Chen, J.L.; Miao, C.P.; Wang, Q.; Zhou, H.; Chen, Y.W.; Li, Y.Q.; Ding, Z.T.; Zhao, L.X. Koninginins N-Q, polyketides from the endophytic fungus Trichoderma koningiopsis Harbored in Panax notoginseng. Nat. Prod. Bioprospect. 2016, 6, 49–55. [Google Scholar] [CrossRef]
- Lang, B.Y.; Li, J.; Zhou, X.X.; Chen, Y.H.; Yang, Y.H.; Li, X.N.; Zeng, Y.; Zhao, P.J. Koninginins L and M, two polyketides from Trichoderma koningii 8662. Phytochem. Lett. 2015, 11, 1–4. [Google Scholar] [CrossRef]
- Zhou, X.X.; Li, J.; Yang, Y.H.; Zeng, Y.; Zhao, P.J. Three new koninginins from Trichoderma neokongii 8722. Phytochem. Lett. 2014, 8, 137–140. [Google Scholar] [CrossRef]
- Shi, X.S.; Wang, D.J.; Li, X.M.; Li, H.L.; Meng, L.H.; Li, X.; Pi, Y.; Zhou, X.W.; Wang, B.G. Antimicrobial polyketides from Trichoderma koningiopsis QA-3, an endophytic fungus obtained from the medicinal plant Artemisia argyi. RSC Adv. 2017, 7, 51335–51342. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, D.K.; Kuang, M.; Peng, W.W.; Chen, Y.; Tan, J.B.; Kang, F.H.; Xu, K.P.; Zou, Z.X. Rhytidhylides A and B, two new phthalide derivatives from the endophytic fungus Rhytidhysteron sp. BZM-9. Molecules 2021, 26, 6092. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lu, X.X.; Huo, L.Q.; Zhang, S.; Chen, Y.; Zou, Z.X.; Tan, H.B. Sesquiterpenes and steroids from an endophytic Eutypella scoparia. J. Nat. Prod. 2021, 84, 1715–1724. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.W.; Kuang, M.; Huang, Y.T.; Li, M.F.; Zheng, Y.T.; Xu, L.; Tan, J.B.; Kang, F.H.; Tan, H.B.; Zou, Z.X. Pseudocercones A-C, three new polyketide derivatives from the endophytic fungus Pseudocercospora sp. TSS-1. Nat. Prod. Res. 2022, 1–8. [Google Scholar] [CrossRef]
- Kuang, M.; Peng, W.W.; Huang, Y.T.; Li, M.F.; Qin, S.Y.; Zheng, Y.T.; Xu, L.; Huang, Q.; Zou, Z.X. Two new chromone derivatives from the rhizosphere soil fungus Ilyonectria robusta. Nat. Prod. Res. 2022, 1–8. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, H.; Ke, X.; Sang, Z.H.; Kuang, M.; Peng, W.W.; Tan, J.B.; Zheng, Y.T.; Zou, Z.X.; Tan, H.B. Five new secondary metabolites from an endophytic fungus Phomopsis sp. SZSJ-7B. Front. Plant Sci. 2022, 13, 1049015. [Google Scholar] [CrossRef]
- Peng, W.W.; Huang, Q.; Ke, X.; Wang, W.X.; Chen, Y.; Sang, Z.H.; Chen, C.; Qin, S.Y.; Zheng, Y.T.; Tan, H.B.; et al. Koningipyridines A and B, two nitrogen-containing polyketides from fungus Trichoderma koningiopsis SC-5. Front. Plant Sci. 2023, 14, 1161420. [Google Scholar]
- Cutler, H.G.; Himmelsbach, D.S.; Yagen, B.; Arrendale, R.F.; Cox, R.H. Koninginin B: A biologically active congener of koninginin A from Trichoderma koningii. J. Agric. Food Chem. 1991, 39, 977–980. [Google Scholar] [CrossRef]
- Ghisalberti, E.L.; Rowland, C.Y. Antifungal metabolites from Trichoderma harzianum. J. Nat. Prod. 1993, 56, 1799–1804. [Google Scholar] [CrossRef]
- Liu, Z.; Yashiroda, Y.; Sun, P.; Ma, H.; Wang, Y.; Li, L.; Yan, F.; Sun, Y. Argenteolides A and B, glycosylated polyketide-peptide hybrid macrolides from an Actinomycete Streptomyces argenteolus. Org. Lett. 2023, 25, 571–575. [Google Scholar] [CrossRef] [PubMed]
- He, Y.C.; Wang, R.L.; Huang, B.; Dai, Q.; Lin, J. Pholiotone A, a new polyketide derivative from Pholiota sp. Nat. Prod. Res. 2020, 34, 1957–1961. [Google Scholar] [CrossRef] [PubMed]
- Cutler, H.G.; Himmelsbach, D.S.; Arrendale, R.F.; Cole, P.D.; Cox, R.H. Koninginin A: A novel plant growth regulator from Trichoderma koningii (Microbiology & Fermentation Industry). Agric. Biol. Chem. 1989, 53, 2605–26011. [Google Scholar]
- Li, M.F.; Li, G.H.; Zhang, K.Q. Non-Volatile Metabolites from Trichoderma spp. Metabolites 2019, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.J.; Cheng, W.; Heydari, H.; Wang, B.; Zhu, K.; Konuklugil, B.; Lin, W.H. Sorbicillinoid-based metabolites from a sponge-derived fungus Trichoderma saturnisporum. Mar. Drugs 2018, 16, 226. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.Z.; Miao, F.P.; Fang, S.T.; Yin, X.L.; Ji, N.Y. Trichorenins A-C, algicidal tetracyclic metabolites from the marine-alga-epiphytic fungus Trichoderma virens Y13-3. J. Nat. Prod. 2018, 81, 1121–1124. [Google Scholar] [CrossRef]
- Lai, C.; Chen, J.; Liu, J.; Tian, D.; Lan, D.; Liu, T.; Wu, B.; Bi, H.; Tang, J. New polyketides from a hydrothermal vent sediment fungus Trichoderma sp. JWM29-10-1 and their antimicrobial effects. Mar. Drugs 2022, 20, 720. [Google Scholar] [CrossRef]
- Hashem, A.H.; Attia, M.S.; Kandil, E.K.; Fawzi, M.M.; Abdelrahman, A.S.; Khader, M.S.; Khodaira, M.A.; Emam, A.E.; Goma, M.A.; Abdelaziz, A.M. Bioactive compounds and biomedical applications of endophytic fungi: A recent review. Microb. Cell Fact. 2023, 22, 107. [Google Scholar] [CrossRef]
- Toppo, P.; Kagatay, L.L.; Gurung, A.; Singla, P.; Chakraborty, R.; Roy, S.; Mathur, P. Endophytic fungi mediates production of bioactive secondary metabolites via modulation of genes involved in key metabolic pathways and their contribution in different biotechnological sector. 3 Biotech. 2023, 13, 191. [Google Scholar] [CrossRef]
- Tarawneh, A.H.; León, F.; Radwan, M.M.; Rosa, L.H.; Cutler, S.J. Secondary metabolites from the fungus Emericella nidulans. Nat. Prod. Commun. 2013, 8, 1285–1288. [Google Scholar] [CrossRef]
- Pracht, P.; Bohle, F.; Grimme, S. Automated exploration of the low-energy chemical space with fast quantum chemical methods. Phys. Chem. Chem. Phys. 2020, 22, 7169–7192. [Google Scholar] [CrossRef] [PubMed]
- Bannwarth, C.; Ehlert, S.; Grimme, S. GFN2-xTB-An accurate and broadly parametrized self-consistent tight-binding quantum chemical method with multipole electrostatics and density-dependent dispersion contributions. J. Chem. Theory Comput. 2019, 15, 1652–1671. [Google Scholar] [CrossRef] [PubMed]
- Tsuzuki, S.; Uchimaru, T. Accuracy of intermolecular interaction energies, particularly those of hetero-atom containing molecules obtained by DFT calculations with Grimme’s D2, D3 and D3BJ dispersion corrections. Phys. Chem. Chem. Phys. 2020, 22, 22508–22519. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16. Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Chai, J.D.; Head-Gordon, M. Long-range corrected hybrid density functionals with damped atom-atom dispersion corrections. Phys. Chem. Chem. Phys. 2008, 10, 6615–6620. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.K.; Wang, W.X. GIAO 13C NMR calculation with sorted training sets improves accuracy and reliability for structural assignation. J. Org. Chem. 2020, 85, 11350–11358. [Google Scholar] [CrossRef]
- Bruhn, T.; Schaumlöffel, A.; Hemberger, Y.; Bringmann, G. SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef]





| No. | 1 a | 2 a | 3 b | |||
|---|---|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | |
| 1 | 194.0 | 194.6 | 199.4 | |||
| 2 | 2.42 (1H, m); 2.58 (1H, m) | 35.3 | 2.71 (1H, m); 2.56 (1H, m) | 22.5 | 2.63 (1H, m); 2.29 (1H, m) | 34.4 |
| 3 | 2.43 (1H, m); 2.36 (1H, m) | 31.1 | 2.46 (1H, m); 1.90 (1H, m) | 29.7 | 2.18 (1H, m); 1.97 (1H, m) | 30.6 |
| 4 | 4.75 (1H, dd, 9.0, 5.4) | 64.0 | 4.14 (1H, d, 4.8) | 71.7 | 4.38 (1H, td, 5.4, 2.4) | 67.1 |
| 5 | 178.5 | 180.6 | 174.9 | |||
| 6 | 115.1 | 111.3 | 113.2 | |||
| 7 | 5.25 (1H, dt, 6.0) | 79.4 | 5.65 (1H, d, 6.0) | 78.3 | 4.35 (1H, t, 2.4) | 65.4 |
| 8 | 5.25 (1H, dt, 6.0) | 89.5 | 5.05 (1H, d, 6.0) | 93.5 | 2.03 (1H, dt 14.4, 2.4); 1.55 (1H, m) | 29.3 |
| 9 | 4.35 (1H, d, 4.2) | 72.0 | 4.16 (1H, d, 4.8) | 74.6 | 4.14 (1H, td, 12.6, 2.4) | 78.2 |
| 10 | 3.66 (1H, m) | 82.1 | 3.43 (1H, td, 7.2) | 78.6 | 3.68 (1H, td, 12.6, 6.0) | 73.6 |
| 11 | 1.67 (2H, m) | 27.7 | 1.70 (1H, m); 1.64 (1H, m) | 27.1 | 1.64 (2H, m) | 33.8 |
| 12 | 1.29 (2H, m) | 26.1 | 1.31 (2H, m) | 26.0 | 1.55 (1H, m); 1.41 (1H, m) | 26.8 |
| 13 | 1.29 (2H, m) | 29.3 | 1.31 (2H, m) | 29.4 | 1.41 (1H, m); 1.33 (1H, m) | 30.6 |
| 14 | 1.29 (2H, m) | 31.7 | 1.31 (2H, m) | 31.6 | 1.33 (2H, m) | 33.2 |
| 15 | 1.29 (2H, m) | 22.6 | 1.31 (2H, m) | 22.7 | 1.33 (2H, m) | 23.8 |
| 16 | 0.87 (3H, t, 6.6) | 14.0 | 0.87 (3H, t, 7.2) | 14.0 | 0.91 (3H, t, 6.6) | 14.6 |
| 17 | 3.64 (1H, m); 3.55 (1H, m) | 65.4 | ||||
| 18 | 1.15 (3H, t, 6.6) | 16.0 | ||||
| No. | Exptl. δ | 1 | |
|---|---|---|---|
| 1 | abs dev c | ||
| 1 | 194.0 | 192.84 | 1.16 |
| 2 | 35.3 | 34.53 | 0.77 |
| 3 | 31.1 | 29.98 | 1.12 |
| 4 | 64.0 | 64.25 | 0.25 |
| 5 | 178.5 | 177.16 | 1.34 |
| 6 | 115.1 | 119.53 | 4.43 |
| 7 | 79.4 | 79.34 | 0.06 |
| 8 | 89.5 | 89.96 | 0.46 |
| 9 | 72.0 | 74.36 | 2.36 |
| 11 | 82.1 | 79.27 | 2.83 |
| 12 | 27.7 | 28.77 | 1.07 |
| 13 | 26.1 | 25.42 | 0.68 |
| 14 | 29.3 | 28.92 | 0.38 |
| 15 | 31.7 | 30.87 | 0.83 |
| 16 | 22.6 | 22.83 | 0.23 |
| MAE a | 1.15 | ||
| RMS b | 1.60 | ||
| Pmean | 30.45% | ||
| Prel | 100% | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, W.; Tan, J.; Sang, Z.; Huang, Y.; Xu, L.; Zheng, Y.; Qin, S.; Tan, H.; Zou, Z. Koninginins X-Z, Three New Polyketides from Trichoderma koningiopsis SC-5. Molecules 2023, 28, 7848. https://doi.org/10.3390/molecules28237848
Peng W, Tan J, Sang Z, Huang Y, Xu L, Zheng Y, Qin S, Tan H, Zou Z. Koninginins X-Z, Three New Polyketides from Trichoderma koningiopsis SC-5. Molecules. 2023; 28(23):7848. https://doi.org/10.3390/molecules28237848
Chicago/Turabian StylePeng, Weiwei, Jianbing Tan, Zihuan Sang, Yuantao Huang, Li Xu, Yuting Zheng, Siyu Qin, Haibo Tan, and Zhenxing Zou. 2023. "Koninginins X-Z, Three New Polyketides from Trichoderma koningiopsis SC-5" Molecules 28, no. 23: 7848. https://doi.org/10.3390/molecules28237848