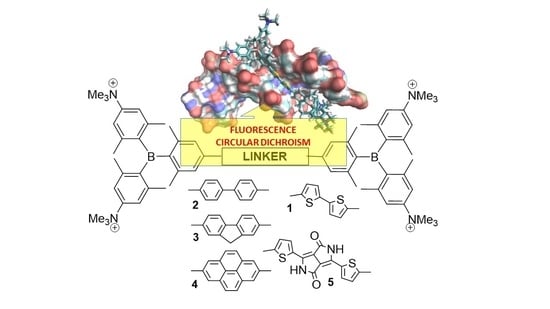
The Nature of the (Oligo/Hetero)Arene Linker Connecting Two Triarylborane Cations Controls Fluorimetric and Circular Dichroism Sensing of Various ds-DNAs and ds-RNAs
Abstract
:1. Introduction
2. Results and Discussion
2.1. Interactions with ds-DNA and ds-RNA
2.1.1. Thermal Denaturation of ds-Polynucleotides
2.1.2. Spectrophotometric Titrations of 2–5 with ds-DNA/RNA
2.1.3. Circular Dichroism
3. Materials and Methods
3.1. General Procedures
3.2. UV/Vis, CD, and Fluorescence Titrations
3.3. Thermal Denaturation Experiments
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Radić Stojković, M.; Škugor, M.; Tomić, S.; Grabar, M.; Smrečki, V.; Dudek, Ł.; Grolik, J.; Eilmes, J.; Piantanida, I. A short, rigid linker between pyrene and guanidiniocarbonyl-pyrrole induced a new set of spectroscopic responses to the ds-DNA secondary structure. Org. Biomol. Chem. 2013, 11, 4077–4085. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Griesbeck, S.; Marder, T.B. Recent developments in and perspectives on three-coordinate boron materials: A bright future. Chem. Sci. 2017, 8, 846–863. [Google Scholar] [CrossRef] [PubMed]
- Entwistle, C.D.; Marder, T.B. Boron Chemistry Lights the Way: Optical Properties of Molecular and Polymeric Systems. Angew. Chem. Int. Ed. Engl. 2002, 41, 2927–2931. [Google Scholar] [CrossRef]
- Entwistle, C.D.; Marder, T.B. Applications of Three-Coordinate Organoboron Compounds and Polymers in Optoelectronics, Special Issue on Organic Electronics. Chem. Mater. 2004, 16, 4574–4585. [Google Scholar] [CrossRef]
- Griesbeck, S.; Zhang, Z.; Gutmann, M.; Lühmann, T.; Edkins, R.M.; Clermont, G.; Lazar, A.N.; Haehnel, M.; Edkins, K.; Eichhorn, A.; et al. Water-Soluble Triarylborane Chromophores for One- and Two-Photon Excited Fluorescence Imaging of Mitochondria in Cells. Chem. Eur. J. 2016, 22, 14701–14706. [Google Scholar] [CrossRef]
- Berger, S.M.; Marder, T.B. Applications of Triarylborane Materials in Cell Imaging and Sensing of Bio-relevant Molecules such as DNA, RNA, and Proteins. Mater. Horiz. 2022, 9, 112–120. [Google Scholar]
- Berger, S.M.; Rühe, J.; Schwarzmann, J.; Phillipps, A.; Richard, A.-K.; Ferger, M.; Krummenacher, I.; Tumir, L.-M.; Ban, Ž.; Crnolatac, I.; et al. Bithiophene-Cored, mono-, bis-, and tris-(Trimethylammonium)-Substituted, bis-Triarylborane Chromophores: Effect of the Number and Position of Charges on Cell Imaging and DNA/RNA Sensing. Chem. Eur. J. 2021, 27, 14057–14072. [Google Scholar] [CrossRef]
- Griesbeck, S.; Ferger, M.; Czernetzi, C.; Wang, C.; Bertermann, R.; Friedrich, A.; Haehnel, M.; Sieh, D.; Taki, M.; Yamaguchi, S.; et al. Optimization of Aqueous Stability versus π-Conjugation in Tetracationic Bis(triarylborane) Chromophores: Applications in Live-Cell Imaging. Chem. Eur. J. 2019, 25, 7679–7688. [Google Scholar] [CrossRef]
- Griesbeck, S.; Michail, E.; Rauch, F.; Ogasawara, H.; Wang, C.; Sato, Y.; Edkins, R.; Zhang, Z.; Taki, M.; Lambert, C.; et al. The Effect of Branching on One- and Two-Photon Absorption, Cell Viability and Localization of Cationic Triarylborane Chromophores with Dipolar vs. Octupolar Charge Distributions for Cellular Imaging. Chem. Eur. J. 2019, 25, 13164–13175. [Google Scholar] [CrossRef]
- Ban, Ž.; Griesbeck, S.; Tomić, S.; Nitsch, J.; Marder, T.B.; Piantanida, I. A Quadrupolar Bis-Triarylborane Chromophore as a Fluorimetric and Chirooptic Probe for Simultaneous and Selective Sensing of DNA, RNA and Proteins. Chem. Eur. J. 2020, 26, 2195–2203. [Google Scholar] [CrossRef]
- Amini, H.; Ban, Ž.; Ferger, M.; Lorenzen, S.; Rauch, F.; Friedrich, A.; Crnolatac, I.; Kenđel, A.; Miljanić, S.; Piantanida, I.; et al. Tetracationic Bis-Triarylborane 1,3-Butadiyne as a Combined Fluorimetric and Raman Probe for Simultaneous and Selective Sensing of Various DNA, RNA, and Proteins. Chem. Eur. J. 2020, 26, 6017–6028. [Google Scholar] [CrossRef] [PubMed]
- Ferger, M.; Roger, C.; Köster, E.; Rauch, F.; Lorenzen, S.; Krummenacher, I.; Friedrich, A.; Košćak, M.; Nestić, D.; Braunschweig, H.; et al. Electron-Rich EDOT Linkers in Tetracationic bis-Triarylborane Chromophores: Influence on Water-Stability, Bio-macromolecule Sensing, and Photoinduced Cytotoxicity. Chem. Eur. J. 2022, 28, e202201130. [Google Scholar] [CrossRef] [PubMed]
- Ferger, M.; Ban, Ž.; Krošl, I.; Tomić, S.; Dietrich, L.; Lorenzen, S.; Rauch, F.; Sieh, D.; Friedrich, A.; Griesbeck, S.; et al. Bis(phenylethynyl)arene Linkers in Tetracationic Bis-triarylborane Chromophores Control Fluorimetric and Raman Sensing of Various DNAs and RNAs. Chem. Eur. J. 2021, 27, 5142–5159. [Google Scholar] [CrossRef] [PubMed]
- Božinović, K.; Nestić, D.; Lambert, C.; Michail, E.; Majhen, D.; Ferger, M.; Marder, T.B.; Košćak, M.; Piantanida, I. Diethynylarene-linked bis(triarylborane)cations as theranostic agents for tumor cell and virus-targeted photodynamic therapy. J. Photochem. Photobiol. B Biol. 2022, 234, 112523. [Google Scholar] [CrossRef]
- Griesbeck, S.; Michail, E.; Wang, C.; Ogasawara, H.; Lorenzen, S.; Gerstner, L.; Zang, T.; Nitsch, J.; Sato, Y.; Bertermann, R.; et al. Tuning the π-bridge of quadrupolar triarylborane chromophores for one- and two-photon excited fluorescence imaging of lysosomes in live cells. Chem. Sci. 2019, 10, 5405–5422. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Lacroix, L. Analysis of Thermal Melting Curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef]
- Scatchard, G. The Attractions of Proteins for Small Molecules and Ions. Ann. N.Y. Acad. Sci. 1949, 51, 660–672. [Google Scholar] [CrossRef]
- McGhee, J.D.; von Hippel, P.H. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef]
- Demeunynck, M.; Demeunynck, M.; Bailly, C.; Wilson, W.D. Small Molecule DNA and RNA Binders: From Synthesis to Nucleic Acid Complexes; Demeunynck, M., Bailly, C., Wilson, W.D., Eds.; Wiley-VCH: Weinheim, Germany, 2003. [Google Scholar]
- Tumir, L.M.; Crnolatac, I.; Deligeorgiev, T.; Vasilev, A.; Kaloyanova, S.; Branilovic, M.G.; Tomic, S.; Piantanida, I. Kinetic Differentiation between Homo- and Alternating AT DNA by Sterically Restricted Phosphonium Dyes. Chem. Eur. J. 2012, 18, 3859–3864. [Google Scholar] [CrossRef]
- Steenken, S.; Jovanovic, S.V. How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. J. Am. Chem. Soc. 1997, 119, 617–618. [Google Scholar] [CrossRef]
- Piantanida, I.; Palm, B.S.; Žinić, M.; Schneider, H.J. A new 4,9-diazapyrenium intercalator for single- and double-stranded nucleic acids: Distinct differences from related diazapyrenium compounds and ethidium bromide. J. Chem. Soc. Perk. Trans. 2001, 2, 1808–1816. [Google Scholar] [CrossRef]
- Rodger, A.; Nordén, B. Circular Dichroism and Linear Dichroism; Oxford University Press: New York, NY, USA, 1997; Chapter 2. [Google Scholar]
- Eriksson, M.; Nordén, B. Linear and circular dichroism of drug-nucleic acid complexes. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2001; Volume 340, pp. 68–98. [Google Scholar]
- Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Polarization spectroscopy methods in the determination of interactions of small molecules with nucleic acids—Tutorial. Beil. J. Org. Chem. 2018, 14, 84–105. [Google Scholar] [CrossRef]
- Chaires, J.B.; Dattagupta, N.; Crothers, D.M. Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: Equilibrium binding studies on interaction of daunomycin with deoxyribonucleic acid. Biochemistry 1982, 21, 3933–3940. [Google Scholar] [CrossRef] [PubMed]
- Chalikian, T.V.; Völker, J.; Plum, G.E.; Breslauer, K.J. A more unified picture for the thermodynamics of nucleic acid duplex melting: A characterization by calorimetric and volumetric techniques. Proc. Natl. Acad. Sci. USA 1999, 96, 7853–7858. [Google Scholar] [CrossRef] [PubMed]
- Tumir, L.M.; Piantanida, I.; Juranovic, I.; Meic, Z.; Tomic, S.; Zinic, M. Recognition of homo-polynucleotides containing adenine by a phenanthridinium bis-uracil conjugate in aqueous media. Chem. Commun. 2005, 20, 2561–2563. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Qi, Z.W.; Wu, Y.L.; Zhao, J.; Wang, L.; Peng, D.F.; Zhang, Y.; Huang, X.A.; Liu, F. A photosensitizing perylenediimide dye lights up cell nucleolus through visible light-mediated intracellular translocation. Dye. Pigment. 2021, 196, 109722. [Google Scholar] [CrossRef]
- Cesaretti, A.; Calzoni, E.; Montegiove, N.; Bianconi, T.; Alebardi, M.; La Serra, M.A.; Consiglio, G.; Fortuna, C.G.; Elisei, F.; Spalletti, A. Lighting-Up the Far-Red Fluorescence of RNA-Selective Dyes by Switching from Ortho to Para Position. Int. J. Mol. Sci. 2023, 24, 4812. [Google Scholar] [CrossRef]
- Deng, K.L.; Wang, L.; Xia, Q.; Liu, R.Y.; Qu, J.Q. A nucleic acid-specific fluorescent probe for nucleolus imaging in living cells. Talanta 2019, 192, 212–219. [Google Scholar] [CrossRef]
- Saenger, W. Principles of Nucleic Acid Structure; Springer: New York, NY, USA, 1983; p. 226. [Google Scholar]
- Cantor, C.R.; Schimmel, P.R. Biophysical Chemistry Part III: The Behavior of Biological Macromolecules; W.H. Freeman and Company: San Francisco, CA, USA, 1980; pp. 1109–1181. [Google Scholar]







| ct-DNA | p(dAdT)2 | poly rA—poly rU | |
|---|---|---|---|
| c 1 | 7.3 | 10.0 | 9.5 |
| 2 | 4.5 | 6.7 | 12.1 |
| 3 | 10.0 | 8.2 | 9.3 |
| 4 | 8.0 | 9.6 | 7.4 |
| 5 | 3.4 | - | 0.6 |
| 1 [10] | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|
| ct-DNA | 7.0 | b 6.1 | 6.9 | 7.1 | 7.7 |
| p(dAdT)2 | 7.9 | b 6.1 | 6.9 | 8.7 | 7.7 |
| p(dGdC)2 | 7.6 | b 6.0 | 7.4 | 8.0 | 7.9 |
| prAprU | b 7 | b 7.5 | c 7.9 | 7.4 | 7.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tumir, L.-M.; Pavlović Saftić, D.; Crnolatac, I.; Ban, Ž.; Maslać, M.; Griesbeck, S.; Marder, T.B.; Piantanida, I. The Nature of the (Oligo/Hetero)Arene Linker Connecting Two Triarylborane Cations Controls Fluorimetric and Circular Dichroism Sensing of Various ds-DNAs and ds-RNAs. Molecules 2023, 28, 4348. https://doi.org/10.3390/molecules28114348
Tumir L-M, Pavlović Saftić D, Crnolatac I, Ban Ž, Maslać M, Griesbeck S, Marder TB, Piantanida I. The Nature of the (Oligo/Hetero)Arene Linker Connecting Two Triarylborane Cations Controls Fluorimetric and Circular Dichroism Sensing of Various ds-DNAs and ds-RNAs. Molecules. 2023; 28(11):4348. https://doi.org/10.3390/molecules28114348
Chicago/Turabian StyleTumir, Lidija-Marija, Dijana Pavlović Saftić, Ivo Crnolatac, Željka Ban, Matea Maslać, Stefanie Griesbeck, Todd B. Marder, and Ivo Piantanida. 2023. "The Nature of the (Oligo/Hetero)Arene Linker Connecting Two Triarylborane Cations Controls Fluorimetric and Circular Dichroism Sensing of Various ds-DNAs and ds-RNAs" Molecules 28, no. 11: 4348. https://doi.org/10.3390/molecules28114348