Novel Disulfiram Derivatives as ALDH1a1-Selective Inhibitors
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Chemical Synthesis of Compounds
3.2. ALDH Enzyme Inhibition Assay
3.3. Molecular Modelling
4. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Poturnajova, M.; Kozovska, Z.; Matuskova, M. Aldehyde dehydrogenase 1A1 and 1A3 isoforms-mechanism of activation and regulation in cancer. Cell Signal 2021, 87, 110120. [Google Scholar] [CrossRef]
- Jackson, B.; Brocker, C.; Thompson, D.C.; Black, W.; Vasiliou, K.; Nebert, D.W.; Vasiliou, V. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum. Genom. 2011, 5, 283–303. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Brocker, C.; Koppaka, V.; Chen, Y.; Jackson, B.C.; Matsumoto, A.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol. Med. 2013, 56, 89–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasiliou, V.; Pappa, A.; Estey, T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab. Rev. 2004, 36, 279–299. [Google Scholar] [CrossRef] [PubMed]
- Pierre-Louis, O.; Clay, D.; Brunet de la Grange, P.; Blazsek, I.; Desterke, C.; Guerton, B.; Blondeau, C.; Malfuson, J.V.; Prat, M.; Bennaceur-Griscelli, A.; et al. Dual SP/ALDH functionalities refine the human hematopoietic Lin−CD34+CD38− stem/progenitor cell compartment. Stem. Cells 2009, 27, 2552–2562. [Google Scholar] [CrossRef] [PubMed]
- Reuben, J.M.; Lee, B.N.; Gao, H.; Cohen, E.N.; Mego, M.; Giordano, A.; Wang, X.; Lodhi, A.; Krishnamurthy, S.; Hortobagyi, G.N.; et al. Primary breast cancer patients with high risk clinicopathologic features have high percentages of bone marrow epithelial cells with ALDH activity and CD44+CD24lo cancer stem cell phenotype. Eur. J. Cancer 2011, 47, 1527–1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nwani, N.G.; Condello, S.; Wang, Y.; Swetzig, W.M.; Barber, E.; Hurley, T.; Matei, D. A Novel ALDH1A1 Inhibitor Targets Cells with Stem Cell Characteristics in Ovarian Cancer. Cancers 2019, 11, 502. [Google Scholar] [CrossRef] [Green Version]
- Tanei, T.; Morimoto, K.; Shimazu, K.; Kim, S.J.; Tanji, Y.; Taguchi, T.; Tamaki, Y.; Noguchi, S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin. Cancer Res. 2009, 15, 4234–4241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vassalli, G. Aldehyde Dehydrogenases: Not Just Markers, but Functional Regulators of Stem Cells. Stem. Cells Int. 2019, 2019, 3904645. [Google Scholar] [CrossRef] [Green Version]
- Ziouzenkova, O.; Orasanu, G.; Sharlach, M.; Akiyama, T.E.; Berger, J.P.; Viereck, J.; Hamilton, J.A.; Tang, G.; Dolnikowski, G.G.; Vogel, S.; et al. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat. Med. 2007, 13, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, F.W.; Orasanu, G.; Nallamshetty, S.; Brown, J.D.; Wang, H.; Luger, P.; Qi, N.R.; Burant, C.F.; Duester, G.; Plutzky, J. Retinaldehyde dehydrogenase 1 coordinates hepatic gluconeogenesis and lipid metabolism. Endocrinology 2012, 153, 3089–3099. [Google Scholar] [CrossRef] [Green Version]
- Omran, Z.; Sheikh, R.; Baothman, O.A.; Zamzami, M.A.; Alarjah, M. Repurposing Disulfiram as an Anti-Obesity Drug: Treating and Preventing Obesity in High-Fat-Fed Rats. Diabetes Metab. Syndr. Obes. 2020, 13, 1473–1480. [Google Scholar] [CrossRef] [PubMed]
- Sanders, T.J.; McCarthy, N.E.; Giles, E.M.; Davidson, K.L.; Haltalli, M.L.; Hazell, S.; Lindsay, J.O.; Stagg, A.J. Increased production of retinoic acid by intestinal macrophages contributes to their inflammatory phenotype in patients with Crohn’s disease. Gastroenterology 2014, 146, 1278–1288.e2. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Molotkov, A.; Manabe, S.; Donmoyer, C.M.; Deltour, L.; Foglio, M.H.; Cuenca, A.E.; Blaner, W.S.; Lipton, S.A.; Duester, G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol. Cell. Biol. 2003, 23, 4637–4648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Yang, K.; Liang, D.; Jiang, C.; Ma, Z. Discovery and development of selective aldehyde dehydrogenase 1A1 (ALDH1A1) inhibitors. Eur. J. Med. Chem. 2021, 209, 112940. [Google Scholar] [CrossRef]
- Lipsky, J.J.; Shen, M.L.; Naylor, S. In vivo inhibition of aldehyde dehydrogenase by disulfiram. Chem. Biol. Interact 2001, 130–132, 93–102. [Google Scholar] [CrossRef]
- Yang, Q.; Yao, Y.; Li, K.; Jiao, L.; Zhu, J.; Ni, C.; Li, M.; Dou, Q.P.; Yang, H. An Updated Review of Disulfiram: Molecular Targets and Strategies for Cancer Treatment. Curr. Pharm. Des. 2019, 25, 3248–3256. [Google Scholar] [CrossRef]
- Moore, S.A.; Baker, H.M.; Blythe, T.J.; Kitson, K.E.; Kitson, T.M.; Baker, E.N. Sheep liver cytosolic aldehyde dehydrogenase: The structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure 1998, 6, 1541–1551. [Google Scholar] [CrossRef] [Green Version]
- Omran, Z. Development of new disulfiram analogues as ALDH1a1-selective inhibitors. Bioorganic Med. Chem. Lett. 2021, 40, 127958. [Google Scholar] [CrossRef]
- Omran, Z. New Disulfiram Derivatives as MAGL-Selective Inhibitors. Molecules 2021, 26, 3296. [Google Scholar] [CrossRef]
- Buchman, C.D.; Hurley, T.D. Inhibition of the Aldehyde Dehydrogenase 1/2 Family by Psoralen and Coumarin Derivatives. J. Med. Chem. 2017, 60, 2439–2455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef] [PubMed]





| Compound | R | IC50 (µM) ± SE (ALDH1a1) | IC50 (µM) ± SE (ALDH2) |
|---|---|---|---|
| 1 (Disulfiram) | Et | 0.15 ± 0.01 | 3.85 ± 0.10 |
| 2 |  | 0.17 ± 0.06 | NI |
| 4a |  | 0.31 ± 0.14 | NI |
| 4b |  | 0.59 ± 0.46 | NI |
| 4c |  | 0.86 ± 0.68 | NI |
| 4d |  | NI | NI |
| 4e |  | 0.58 ± 0.41 | NI |
| 4f |  | 5.76 ± 4.01 | NI |
| 4g |  | 0.39 ± 0.27 | 200.22 ± 129.08 |
| 4h |  | 0.39 ± 0.31 | NI |
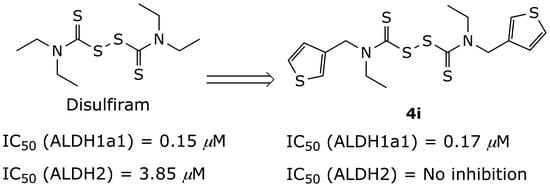
| 4i |  | 0.17 ± 0.10 | NI |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omran, Z. Novel Disulfiram Derivatives as ALDH1a1-Selective Inhibitors. Molecules 2022, 27, 480. https://doi.org/10.3390/molecules27020480
Omran Z. Novel Disulfiram Derivatives as ALDH1a1-Selective Inhibitors. Molecules. 2022; 27(2):480. https://doi.org/10.3390/molecules27020480
Chicago/Turabian StyleOmran, Ziad. 2022. "Novel Disulfiram Derivatives as ALDH1a1-Selective Inhibitors" Molecules 27, no. 2: 480. https://doi.org/10.3390/molecules27020480