Ruthenium(II)/(III) DMSO-Based Complexes of 2-Aminophenyl Benzimidazole with In Vitro and In Vivo Anticancer Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterization
2.1.1. Synthesis
2.1.2. FTIR Spectra
2.1.3. NMR Spectra
2.1.4. Electronic Spectra and Magnetic Measurements
2.1.5. Density Functional Theory Calculations
2.1.6. Solution Stability
2.1.7. Cyclic Voltammetry
2.2. Biological Studies
2.2.1. UV-Vis Studies of DNA Interactions
2.2.2. Fluorescence studies of DNA interactions
2.3. Anticancer Activity
2.3.1. In Vitro Assays
Effect of Hapbim Ligand and Its Complexes (1) and (2) on MCF7, Caco2, and THLE-Cell Viability
Compound (2) Inhibits Proliferation of MCF7 and Caco2 Cells via Induction of Apoptosis
Effect of Compound (2) on Cell-Cycle Arrest in MCF7 and Caco2 Cells
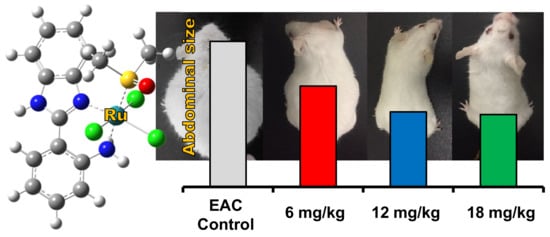
Effect of Compound (2) on Ehrlich Carcinoma Cell Viability
2.3.2. In Vivo Experiments
Antioxidant Activity Assay
Effect of Complex (2) on the Expression of Apoptosis-Related Genes in the Liver of Ehrlich Mice
3. Materials and Methods
3.1. Materials
3.2. Measurements
3.3. Synthesis
3.3.1. [Ru(II)Cl2(DMSO)2(Hapbim)] (1)
3.3.2. [Ru(III)Cl3(DMSO)(Hapbim)]·H2O (2)
3.4. Solution Stability
3.5. Biological Studies
3.5.1. DNA Interactions
UV-Visible Spectra
Fluorescence Spectra
3.6. In Vitro Assays
3.6.1. Cell Viability by MTT Assay
3.6.2. EAC Cytotoxicity Assay (In Vitro)
3.6.3. Detection of DNA Fragmentation
3.6.4. Cell-Cycle Analysis Using Flow Cytometry
3.7. In Vivo Assays
3.7.1. Animal Experiments
3.7.2. Median Lethal Dose (LD50) Determination
3.7.3. In Vivo Tumor Cell Transplantation
- Control group: Mice were intraperitoneally injected with a vehicle (2% DMSO + sterile saline) for 12 days.
- I, II, and III control groups: Healthy normal mice were intraperitoneally injected with 6, 12, and 18 mg/kg of complex (2), respectively, for 12 days.
- EAC group: Mice were intraperitoneally injected by a single dose of 2.5 × 106 viable Ehrlich ascites carcinoma (EAC) cells in PBS. In brief, EAC cells were withdrawn from the EAC mouse’s intraperitoneal cavity, washed, and centrifuged three times with sterile saline. Then, the pellets were suspended in sterile PBS and counted by the trypan blue exclusion assay, which confirmed that the viability of the cells was 98%.
- I, II, and III treated groups: Firstly, 30 healthy mice were intraperitoneally injected with a single dose of 2.5 × 106 viable EAC cells. After one day; the mice were divided into I, II, and III treated groups (10 mice each group), intraperitoneally injected with 6, 12, and 18 mg/kg Ru(III) complex, respectively, for 12 days.
3.7.4. Biochemical Parameters
3.7.5. Oxidative Stress Markers
3.7.6. Molecular Analysis by Real-Time PCR
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Giaccone, G. Clinical perspectives on platinum resistance. Drugs 2000, 59, 9–17. [Google Scholar] [CrossRef]
- Dabrowiak, J.C. Metals in Medicine, 2nd ed.; Wiley: Hoboken, NJ, USA, 2017. [Google Scholar]
- Jaouen, G.; Vessieres, A.; Top, S. Ferrocifen type anti cancer drugs. Chem. Soc. Rev. 2015, 44, 8802–8817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gianferrara, T.; Bratsos, I.; Alessio, E. A categorization of metal anticancer compounds based on their mode of action. Dalton Trans. 2009, 37, 7588–7598. [Google Scholar] [CrossRef] [PubMed]
- Biersack, B.; Ahmad, A.; Sarkar, F.H.; Schobert, R. Coinage metal complexes against breast cancer. Curr. Med. Chem. 2012, 19, 3949–3956. [Google Scholar] [CrossRef] [PubMed]
- Levina, A.; Mitra, A.; Lay, P.A. Recent developments in ruthenium anticancer drugs. Metallomics 2009, 1, 458–470. [Google Scholar] [CrossRef]
- Bergamo, A.; Sava, G. Ruthenium anticancer compounds: Myths and realities of the emerging metal-based drugs. Dalton Trans. 2011, 40, 7817–7823. [Google Scholar] [CrossRef]
- Alessio, E.; Messori, L. NAMI-A and KP1019/1339, Two Iconic Ruthenium Anticancer Drug Candidates Face-to-Face: A Case Story in Medicinal Inorganic Chemistry. Molecules 2019, 24, 1995. [Google Scholar] [CrossRef] [Green Version]
- Coverdale, J.P.C.; Laroiya-McCarron, T.; Romero-Canelón, I. Designing Ruthenium Anticancer Drugs: What Have We Learnt from the Key Drug Candidates? Inorganics 2019, 7, 31. [Google Scholar] [CrossRef] [Green Version]
- Bonetti, A.; Leone, R.; Muggia, F.; Howell, S.B. Platinum and Other Heavy Metal Compounds in Cancer Chemotherapy: Molecular Mechanisms and Clinical Applications; Humana Press, c/o Springer Science+Business Media, LLC: New York, NY, USA, 2009. [Google Scholar]
- Han Ang, W.; Dyson, P.J. Classical and Non-Classical Ruthenium-Based Anticancer Drugs: Towards Targeted Chemotherapy. Eur. J. Inorg. Chem. 2006, 2006, 4003–4018. [Google Scholar] [CrossRef]
- Chatterjee, B.D.; Mitra, A.; De, G.S. Ruthenium Polyaminocarboxylate Complexes. Platin. Met. Rev. 2006, 50, 2–12. [Google Scholar] [CrossRef]
- Sgouros, G.; Emadi, A. Ruthenium-based chemotherapeutics: Are they ready for prime time? Cancer Chemother. Pharmacol. 2010, 66, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Bergamo, A.; Sava, G. Ruthenium complexes can target determinants of tumour malignancy. Dalton Trans. 2007, 13, 1267–1272. [Google Scholar] [CrossRef]
- Hartinger, C.G.; Jakupec, M.A.; Zorbas-Seifried, S.; Groessl, M.; Egger, A.; Berger, W.; Zorbas, H.; Dyson, P.J.; Keppler, B.K. KP1019, a new redox-active anticancer agent—Preclinical development and results of a clinical phase I study in tumor patients. Chem. Biodivers 2008, 5, 2140–2155. [Google Scholar] [CrossRef] [PubMed]
- Trondl, R.; Heffeter, P.; Kowol, C.; Jakupec, M.A.; Berger, W.; Keppler, B.K. NKP-1339, the first ruthenium-based anticancer drug on the edge to clinical application. Chem. Sci. 2014, 5, 2925–2932. [Google Scholar] [CrossRef] [Green Version]
- Hartinger, C.G.; Zorbas-Seifried, S.; Jakupec, M.A.; Kynast, B.; Zorbas, H.; Keppler, B.K. From bench to bedside-preclinical and early clinical development of the anticancer agent indazolium trans-[tetrachlorobis(1H-indazole)ruthenate(III)] (KP1019 or FFC14A). J. Inorg. Biochem. 2006, 100, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Lentz, F.; Drescher, A.; Lindauer, A.; Henke, M.; Hilger, R.A.; Hartinger, C.G.; Scheulen, M.E.; Dittrich, C.; Keppler, B.K.; Jaehde, U. Pharmacokinetics of a novel anticancer ruthenium complex (KP1019, FFC14A) in a phase I dose-escalation study. Anti-Cancer Drugs 2009, 20, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Alessio, E.; Messori, L. The deceptively similar ruthenium(III) drug candidates KP1019 and NAMI-A have different actions. what did we learn in the past 30 years? In Metallo-Drugs: Development and Action of Anticancer Agents; Sigel, A., Freisinger, E., Sigel, R.K.O., Eds.; Walter de Gruyter GmbH: Berlin, Germany; Boston, MA, USA, 2018; p. 141. [Google Scholar]
- Lazarevic, T.; Rilak, A.; Bugarčić, Ž.D. Platinum, palladium, gold and ruthenium complexes as anticancer agents: Current clinical uses, cytotoxicity studies and future perspectives. Eur. J. Med. Chem. 2017, 142, 8–31. [Google Scholar] [CrossRef]
- Mu, C.; Walsby, C.J. Ruthenium Anticancer Compounds with Biologically-derived Ligands. In Ligand Design in Medicinal Inorganic Chemistry; Storr, T., Ed.; John Wiley & Sons Inc.: Chichester, UK, 2014; pp. 405–437. [Google Scholar]
- Alessio, E. Thirty Years of the Drug Candidate NAMI-A and the Myths in the Field of Ruthenium Anticancer Compounds: A Personal Perspective. Eur. J. Inorg. Chem. 2016, 2017, 1549–1560. [Google Scholar] [CrossRef]
- Małecki, J.G. Half-sandwich ruthenium(II) complexes with N- and N,(N,O)-donor ligands: Molecular, electronic structures, and computational study. Struct. Chem. 2011, 23, 461–472. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Kumar, A.; Gupta, R.K.; Paitandi, R.P.; Singh, K.B.; Trigun, S.K.; Hundal, M.S.; Pandey, D.S. Cationic Ru(II), Rh(III) and Ir(III) complexes containing cyclic π-perimeter and 2-aminophenyl benzimidazole ligands: Synthesis, molecular structure, DNA and protein binding, cytotoxicity and anticancer activity. J. Organomet. Chem. 2016, 801, 68–79. [Google Scholar] [CrossRef]
- Paul, A.; Anbu, S.; Sharma, G.; Kuznetsov, M.L.; Koch, B.; Da Silva, M.F.C.G.; Pombeiro, A.J.L. Synthesis, DNA binding, cellular DNA lesion and cytotoxicity of a series of new benzimidazole-based Schiff base copper(II) complexes. Dalton Trans. 2015, 44, 19983–19996. [Google Scholar] [CrossRef] [PubMed]
- Yellol, J.; Pérez, S.A.; Buceta, A.; Yellol, G.; Donaire, A.; Szumlas, P.; Bednarski, P.J.; Makhloufi, G.; Janiak, C.; Espinosa, A.; et al. Novel C,N-Cyclometalated Benzimidazole Ruthenium(II) and Iridium(III) Complexes as Antitumor and Antiangiogenic Agents: A Structure–Activity Relationship Study. J. Med. Chem. 2015, 58, 7310–7327. [Google Scholar] [CrossRef] [PubMed]
- Prosser, K.E.; Chang, S.W.; Saraci, F.; Le, P.H.; Walsby, C.J. Anticancer copper pyridine benzimidazole complexes: ROS generation, biomolecule interactions, and cytotoxicity. J. Inorg. Biochem. 2017, 167, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, L.; Raj, V.V.; Raghunandan, N.; Venkateshwerlu, L. ChemInform Abstract: Recent Advances and Potential Pharmacological Activities of Benzimidazole Derivatives. Pharma Chem. 2011, 3, 172–193. [Google Scholar] [CrossRef]
- Sahyon, H.A.; El-Bindary, A.A.; Shoair, A.; Abdellatif, A. Synthesis and characterization of ruthenium(III) complex containing 2-aminomethyl benzimidazole, and its anticancer activity of in vitro and in vivo models. J. Mol. Liq. 2018, 255, 122–134. [Google Scholar] [CrossRef]
- Sava, G.; Pacor, S.; Bregant, F.; Ceschia, V.; Mestroni, G. Metal complexes of ruthenium. Anti-Cancer Drugs 1990, 1, 99–108. [Google Scholar] [CrossRef]
- Biradar, N.; Goudar, T. Addition compounds of niobium(V) with 2-substituted benzimidazoles. J. Inorg. Nucl. Chem. 1977, 39, 358–360. [Google Scholar] [CrossRef]
- Tamayo, P.; Mendiola, M.A.; Masaguer, J.R.; Molleda, C. Tin(IV), titanium(IV) and vanadium(IV) chloride complexes with 2-aminobenzimidazole and 2 (2′-aminophenyl) benzimidazole. Transit. Met. Chem. 1989, 14, 283–286. [Google Scholar] [CrossRef]
- Elsayed, S.A.; Gaml, E.A.; Nasher, M. New ruthenium(II) bipyridine complex bearing 2-aminophenylbenzimidazole: Synthesis, spectral characterization and optical properties. Opt. Mater. 2018, 84, 8–15. [Google Scholar] [CrossRef]
- Evans, I.P.; Spencer, A.; Wilkinson, G. Dichlorotetrakis(dimethyl sulphoxide)ruthenium(II) and its use as a source material for some new ruthenium(II) complexes. J. Chem. Soc. Dalton Trans. 1973, 204–219. [Google Scholar] [CrossRef]
- Mahalingam, V.; Chitrapriya, N.; Fronczek, F.R.; Natarajan, K. Dimethyl sulfoxide ruthenium(II) complexes of thiosemicarbazones and semicarbazone: Synthesis, characterization and biological studies. Polyhedron 2008, 27, 2743–2750. [Google Scholar] [CrossRef]
- Mahalingam, V.; Chitrapriya, N.; Fronczek, F.R.; Natarajan, K. New Ru(II)–dmso complexes with heterocyclic hydrazone ligands towards cancer chemotherapy. Polyhedron 2008, 27, 1917–1924. [Google Scholar] [CrossRef]
- Mahalingam, V.; Chitrapriya, N.; Zeller, M.; Natarajan, K. Ru(II)–DMSO complexes containing aromatic and heterocyclic acid hydrazides: Structure, electrochemistry and biological activity. Polyhedron 2009, 28, 1532–1540. [Google Scholar] [CrossRef]
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds; John Wiley and Sons: New York, NY, USA, 1978. [Google Scholar]
- Mahalingam, V.; Chitrapriya, N.; Fronczek, F.R.; Natarajan, K. New Ru(II)-DMSO complexes of ON/SN chelates: Sythesis, behavior of Schiff bases towards hydrolytic cleavage of C=N bond, electrochemistry and biological activities. Polyhedron 2010, 29, 3363–3371. [Google Scholar] [CrossRef]
- Iengo, E.; Mestroni, G.; Geremia, S.; Calligaris, M.; Alessio, E. Novel ruthenium(III) dimers Na2[{trans-RuCl4(Me2SO-S)}2(µ-L)] and [{mer,cis-RuCl3(Me2SO-S)(Me2SO-O)}2 (µ-L)](L= bridging heterocyclic N-donor ligand) closely related to the antimetastatic complex Na[trans-RuCl4(Me2SO-S)(Him)]. J. Chem. Soc. Dalton Trans. 1999, 3361–3371. [Google Scholar] [CrossRef]
- Lever, A.B.P.; Dodsworth, E.S. Electrochemistry, charge transfer spectroscopy, and electronic structure. In Inorganic Electronic Structure and Spectroscopy; Solomon, E.I., Lever, A.B.P., Eds.; Wiley-Interscience: New York, NY, USA, 1999; Volume 2, pp. 227–289. [Google Scholar]
- Taqui Khan, M.M.; Srinivas, D.; Kureshy, R.I.; Khan, N.H. Synthesis, characterization, and EPR studies of stable ruthenium(III) Schiff base chloro and carbonyl complexes. Inorg. Chem. 1990, 29, 2320–2326. [Google Scholar] [CrossRef]
- Singh, M.K.; Kar, N.K.; Lal, R.A.; Asthana, M. Synthesis and spectral studies on heterobimetallic complexes of manganese and ruthenium derived from N-(2-hydroxynaphthalen-1-yl)methylenebenzoylhydrazide. J. Coord. Chem. 2009, 62, 2893–2902. [Google Scholar] [CrossRef]
- Frisch, M.J.T.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision E.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Meier, S.M.; Gerner, C.; Berger, W.; Hartinger, C.G.; Keppler, B.K. Structure–activity relationships for ruthenium and osmium anticancer agents–towards clinical development. Chem. Soc. Rev. 2018, 47, 909–928. [Google Scholar] [CrossRef]
- Sava, G.; Pacor, S.; Zorzet, S.; Alessio, E.; Mestroni, G. Antitumour properties of dimethylsulphoxide ruthenium(II) complexes in the Lewis lung carcinoma system. Pharmacol. Res. 1989, 21, 617–628. [Google Scholar] [CrossRef]
- Bratsos, I.; Bergamo, A.; Sava, G.; Gianferrara, T.; Zangrando, E.; Alessio, E. Influence of the anionic ligands on the anticancer activity of Ru(II)–dmso complexes: Kinetics of aquation and in vitro cytotoxicity of new dicarboxylate compounds in comparison with their chloride precursors. J. Inorg. Biochem. 2008, 102, 606–617. [Google Scholar] [CrossRef]
- Alagesan, M.; Sathyadevi, P.; Krishnamoorthy, P.; Bhuvanesh, N.S.P.; Dharmaraj, N. DMSO containing ruthenium(II) hydrazone complexes: In vitro evaluation of biomolecular interaction and anticancer activity. Dalton Trans. 2014, 43, 15829–15840. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.W.; Lewis, A.R.; Prosser, K.E.; Thompson, J.R.; Gladkikh, M.; Bally, M.B.; Warren, J.J.; Walsby, C.J. CF3 Derivatives of the Anticancer Ru(III) Complexes KP1019, NKP-1339, and Their Imidazole and Pyridine Analogues Show Enhanced Lipophilicity, Albumin Interactions, and Cytotoxicity. Inorg. Chem. 2016, 55, 4850–4863. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.-P.; Hu, S.; Liu, J.; Ji, L. Synthesis, characterization, antiproliferative and anti-metastatic properties of two ruthenium–DMSO complexes containing 2,2′-biimidazole. Eur. J. Med. Chem. 2011, 46, 1555–1563. [Google Scholar] [CrossRef] [PubMed]
- El-Hendawy, A.M.; Fayed, A.M.; Mostafa, M.R. Complexes of a diacetylmonoxime Schiff base of S-methyldithiocarbazate (H2damsm) with Fe(III), Ru(III)/Ru(II), and V(IV); catalytic activity and X-ray crystal structure of [Fe(Hdamsm)2]NO3·H2O. Transit. Met. Chem. 2011, 36, 351–361. [Google Scholar] [CrossRef]
- Creutz, C. Complexities of ascorbate as a reducing agent. Inorg. Chem. 1981, 20, 4449–4452. [Google Scholar] [CrossRef]
- Rost, J.; Rapoport, S. Reduction-potential of Glutathione. Nature 1964, 201, 185. [Google Scholar] [CrossRef]
- Schluga, P.; Hartinger, C.G.; Egger, A.; Reisner, E.; Galanski, M.; Jakupec, M.A.; Keppler, B.K. Redox behavior of tumor-inhibiting ruthenium(III) complexes and effects of physiological reductants on their binding to GMP. Dalton Trans. 2006, 1796–1802. [Google Scholar] [CrossRef]
- Jakupec, M.A.; Reisner, E.; Eichinger, A.; Pongratz, M.; Arion, V.B.; Galanski, M.; Hartinger, C.G.; Keppler, B.K. Redox-Active Antineoplastic Ruthenium Complexes with Indazole: Correlation of in Vitro Potency and Reduction Potential. J. Med. Chem. 2005, 48, 2831–2837. [Google Scholar] [CrossRef]
- Long, E.C.; Barton, J.K. On demonstrating DNA intercalation. Acc. Chem. Res. 1990, 23, 271–273. [Google Scholar] [CrossRef]
- Alagesan, M.; Bhuvanesh, N.S.P.; Dharmaraj, N. An investigation on new ruthenium(II) hydrazone complexes as anticancer agents and their interaction with biomolecules. Dalton Trans. 2014, 43, 6087–6099. [Google Scholar] [CrossRef]
- Jiang, C.; Chao, H.; Li, H.; Ji, L. Syntheses, characterization and DNA-binding studies of ruthenium(II) terpyridine complexes: [Ru(tpy)(PHBI)]2+ and [Ru(tpy)(PHNI)]2+. J. Inorg. Biochem. 2003, 93, 247–255. [Google Scholar] [CrossRef]
- Arjmand, F.; Parveen, S.; Afzal, M.; Shahid, M. Synthesis, characterization, biological studies (DNA binding, cleavage, antibacterial and topoisomerase I) and molecular docking of copper(II) benzimidazole complexes. J. Photochem. Photobiol. B Biol. 2012, 114, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Sathyadevi, P.; Krishnamoorthy, P.; Jayanthi, E.; Butorac, R.R.; Cowley, A.H.; Dharmaraj, N. Studies on the effect of metal ions of hydrazone complexes on interaction with nucleic acids, bovine serum albumin and antioxidant properties. Inorg. Chim. Acta 2012, 384, 83–96. [Google Scholar] [CrossRef]
- LePecq, J.B.; Paoletti, C. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J. Mol. Biol. 1967, 27, 87–106. [Google Scholar] [CrossRef]
- Subbaraj, P.; Ramu, A.; Raman, N.; Dharmaraja, J. Synthesis, characterization, DNA interaction and pharmacological studies of substituted benzophenone derived Schiff base metal(II) complexes. J. Saudi Chem. Soc. 2015, 19, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Dimiza, F.; Papadopoulos, A.N.; Tangoulis, V.; Psycharis, V.; Raptopoulou, C.P.; Kessissoglou, D.P.; Psomas, G. Biological evaluation of cobalt(II) complexes with non-steroidal anti-inflammatory drug naproxen. J. Inorg. Biochem. 2012, 107, 54–64. [Google Scholar] [CrossRef]
- Lakshmipraba, J.; Arunachalam, S.; Gandi, D.A.; Thirunalasundari, T. Synthesis, nucleic acid binding and cytotoxicity of polyethyleneimine-copper(II) complexes containing 1,10-phenanthroline and L-valine. Eur. J. Med. Chem. 2011, 46, 3013–3021. [Google Scholar] [CrossRef]
- Sarhan, A.M.; Elsayed, S.A.; Mashaly, M.M.; El-Hendawy, A.M. Oxovanadium(IV) and ruthenium(II) carbonyl complexes of ONS-donor ligands derived from dehydroacetic acid and dithiocarbazate: Synthesis, characterization, antioxidant activity, DNA binding and in vitro cytotoxicity. Appl. Organomet. Chem. 2018, 33, e4655. [Google Scholar] [CrossRef]
- Kostova, I. Ruthenium complexes as anticancer agents. Curr. Med. Chem. 2006, 13, 1085–1107. [Google Scholar] [CrossRef]
- Vajs, J.; Pevec, A.; Gazvoda, M.; Urankar, D.; Goreshnik, E.; Polanc, S.; Košmrlj, J. Synthesis and X-ray Structural Analysis of the Ruthenium(III) Complex Na[trans-RuCl4(DMSO) (PyrDiaz)], the Diazene Derivative of Antitumor NAMI-Pyr. Acta Chim. Slov. 2017, 64, 763–770. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.; Panichpisal, K.; Kurtzman, N.; Nugent, K. Cisplatin Nephrotoxicity: A Review. Am. J. Med. Sci. 2007, 334, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Bunel, V.; Tournay, Y.; Baudoux, T.; De Prez, E.; Marchand, M.; Mekinda, Z.; Maréchal, R.; Roumeguère, T.; Antoine, M.-H.; Nortier, J. Early detection of acute cisplatin nephrotoxicity: Interest of urinary monitoring of proximal tubular biomarkers. Clin. Kidney J. 2017, 10, 639–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergamo, A.; Zorzet, S.; Gava, B.; Sorc, A.; Alessio, E.; Iengo, E.; Sava, G. Effects of NAMI-A and some related ruthenium complexes on cell viability after short exposure of tumor cells. Anti-Cancer Drugs 2000, 11, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Deepa, P.R.; Vandhana, S.; Jayanthi, U.; Krishnakumar, S. Therapeutic and Toxicologic Evaluation of Anti-Lipogenic Agents in Cancer Cells Compared with Non-Neoplastic Cells. Basic Clin. Pharmacol. Toxicol. 2012, 110, 494–503. [Google Scholar] [CrossRef]
- United States Food and Drug Administration (FDA). Guidance for Industry: Safety Testing of Drug Metabolites; Center for Drug Evaluation and Research: Rockville, MD, USA, 2008.
- Goldar, S.; Khaniani, M.S.; Derakhshan, S.M.; Baradaran, B. Molecular Mechanisms of Apoptosis and Roles in Cancer Development and Treatment. Asian Pac. J. Cancer Prev. 2015, 16, 2129–2144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.-H.; Xu, M. DNA fragmentation in apoptosis. Cell Res. 2000, 10, 205–211. [Google Scholar] [CrossRef]
- Pietenpol, J.A.; Stewart, Z.A. Cell cycle checkpoint signaling: Cell cycle arrest versus apoptosis. Toxicology 2002, 181, 475–481. [Google Scholar] [CrossRef]
- El-Magd, M.A.; Khamis, A.; Eldeen, S.K.N.; Ibrahim, W.M.; Salama, A.F. Trehalose enhances the antitumor potential of methotrexate against mice bearing Ehrlich ascites carcinoma. Biomed. Pharmacother. 2017, 92, 870–878. [Google Scholar] [CrossRef]
- Folkman, J. Angiogenesis. Annu. Rev. Med. 2006, 57, 1–18. [Google Scholar] [CrossRef]
- Ellis, L.; Fidler, I. Angiogenesis and metastasis. Eur. J. Cancer 1996, 32, 2451–2460. [Google Scholar] [CrossRef]
- Bielenberg, D.R.; Zetter, B.R. The Contribution of Angiogenesis to the Process of Metastasis. Cancer J. 2015, 21, 267–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; D’Souza, S.S.; Tickoo, S.; Salimath, B.P.; Singh, H. Antiangiogenic and Proapoptotic Activities of Allyl Isothiocyanate Inhibit Ascites Tumor Growth in vivo. Integr. Cancer Ther. 2009, 8, 75–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saraswati, S.; Agrawal, S.; Alhaider, A.A. Ursolic acid inhibits tumor angiogenesis and induces apoptosis through mitochondrial-dependent pathway in Ehrlich ascites carcinoma tumor. Chem. Interact. 2013, 206, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Bhagavan, N.; Ha, C.-E. Clinical Enzymology and Biomarkers of Tissue Injury. In Essentials of Medical Biochemistry; Academic Press: San Diego, CA, USA, 2011; pp. 59–64. [Google Scholar]
- Vadori, M.; Pacor, S.; Vita, F.; Zorzet, S.; Cocchietto, M.; Sava, G. Features and full reversibility of the renal toxicity of the ruthenium-based drug NAMI-A in mice. J. Inorg. Biochem. 2013, 118, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Storz, P. Reactive oxygen species in tumor progression. Front. Biosci. 2005, 10, 1881–1896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dizdaroglu, M.; Jaruga, P. Mechanisms of free radical-induced damage to DNA. Free. Radic. Res. 2012, 46, 382–419. [Google Scholar] [CrossRef] [PubMed]
- Liou, G.-Y.; Storz, P. Reactive oxygen species in cancer. Free. Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef] [Green Version]
- Liochev, S.I.; Fridovich, I. The effects of superoxide dismutase on H2O2 formation. Free. Radic. Biol. Med. 2007, 42, 1465–1469. [Google Scholar] [CrossRef]
- Nagababu, E.; Chrest, F.J.; Rifkind, J.M. Hydrogen-peroxide-induced heme degradation in red blood cells: The protective roles of catalase and glutathione peroxidase. Biochim. Biophys. Acta BBA Gen. Subj. 2003, 1620, 211–217. [Google Scholar] [CrossRef]
- Couto, N.; Wood, J.; Barber, J. The role of glutathione reductase and related enzymes on cellular redox homoeostasis network. Free. Radic. Biol. Med. 2016, 95, 27–42. [Google Scholar] [CrossRef]
- Pinchuk, I.; Shoval, H.; Dotan, Y.; Lichtenberg, D. Evaluation of antioxidants: Scope, limitations and relevance of assays. Chem. Phys. Lipids 2012, 165, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Szatrowski, T.P.; Nathan, C.F. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991, 51, 794–798. [Google Scholar] [PubMed]
- Che, M.-X.; Wang, R.; Li, X.-X.; Wang, H.-Y.; Zheng, X.S. Expanding roles of superoxide dismutases in cell regulation and cancer. Drug Discov. Today 2015, 21, 143–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberley, T.D. Oxidative damage and cancer. Am. J. Pathol. 2002, 160, 403–408. [Google Scholar] [CrossRef] [Green Version]
- Davies, K.P. Oxidative Stress, Antioxidant Defenses, and Damage Removal, Repair, and Replacement Systems. IUBMB Life 2000, 50, 279–289. [Google Scholar] [CrossRef]
- Gulbins, E.; Jekle, A.; Ferlinz, K.; Grassmé, H.; Lang, F. Physiology of apoptosis. Am. J. Physiol. Physiol. 2000, 279, F605–F615. [Google Scholar] [CrossRef]
- Ryter, S.W.; Kim, H.P.; Hoetzel, A.; Park, J.W.; Nakahira, K.; Wang, X.; Choi, A.M.K. Mechanisms of Cell Death in Oxidative Stress. Antioxid. Redox Signal. 2007, 9, 49–89. [Google Scholar] [CrossRef]
- Tsujimoto, Y. Cell death regulation by the Bcl-2 protein family in the mitochondria. J. Cell. Physiol. 2003, 195, 158–167. [Google Scholar] [CrossRef]
- Antonsson, B.; Montessuit, S.; Sanchez, B.; Martinou, J.-C. Bax Is Present as a High Molecular Weight Oligomer/Complex in the Mitochondrial Membrane of Apoptotic Cells. J. Biol. Chem. 2001, 276, 11615–11623. [Google Scholar] [CrossRef] [Green Version]
- Petersen, R.B.; Nunomura, A.; Lee, H.-G.; Casadesus, G.; Perry, G.; Smith, M.A.; Zhu, X. Signal Transduction Cascades Associated with Oxidative Stress in Alzheimer’s Disease. J. Alzheimer’s Dis. 2007, 11, 143–152. [Google Scholar] [CrossRef]
- El-Wahab, S.M.A.; Fouda, F.M. Histological and histochemical study on the effect of Ehrlich ascites carcinoma on the liver and kidney of mice and the possible protective role of tetrodotoxin. Egypt. J. Biol. 2009, 11, 13–25. [Google Scholar]
- Beydogan, A.B.; Bolkent, S. The effects of silibin administration for different time periods on mouse liver with Ehrlich ascites carcinoma. Pharmacol. Rep. 2016, 68, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.I.; Ibrahim, R.Y. Some genetic profiles in liver of Ehrlich ascites tumor-bearing mice under the stress of irradiation. J. Radiat. Res. Appl. Sci. 2014, 7, 188–197. [Google Scholar] [CrossRef]
- Lah, J.J.; Cui, W.; Hu, K.-Q. Effects and mechanisms of silibinin on human hepatoma cell lines. World J. Gastroenterol. 2007, 13, 5299–5305. [Google Scholar] [CrossRef] [PubMed]
- Alessio, E.; Balducci, G.; Calligaris, M.; Costa, G.; Attia, W.M.; Mestroni, G. Synthesis, molecular structure, and chemical behavior of hydrogen trans-bis(dimethyl sulfoxide)tetrachlororuthenate(III) and mer-trichlorotris(dimethyl sulfoxide)ruthenium(III): The first fully characterized chloride-dimethyl sulfoxide-ruthenium(III) complexes. Inorg. Chem. 1991, 30, 609–618. [Google Scholar] [CrossRef]
- Wolfe, A.R.; Shimer, G.H.; Meehan, T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer Science+Business Media: New York, NY, USA, 2013. [Google Scholar]
- Devi, P.U.; Solomon, F.E.; Sharada, A.C. In vivo tumor inhibitory and radiosensitizing effects of an Indian medicinal plant, Plumbago rosea on experimental mouse tumors. Indian J. Exp. Biol. 1994, 32, 523–528. [Google Scholar]
- Chinedu, E.; Arome, D.; Ameh, F.S. A New Method for Determining Acute Toxicity in Animal Models. Toxicol. Int. 2013, 20, 224–226. [Google Scholar] [CrossRef] [Green Version]
- Murrary, R.I. Aspartate aminotransferase. In Clinical Chemistry: Theory, Analysis, and Correlation, 2nd ed.; Kaplan, L.A., Pesce, A.J., Eds.; The CV Mosby Company: St. Louis, MO, USA, 1989; pp. 1112–1116. [Google Scholar]
- Murray, R.L. Alanine aminotransferase. In Clinical Chemistry: Theory, Analysis, and Correlation, 2nd ed.; Kaplan, L.A., Pesce, A.J., Eds.; The CV Mosby Company: St. Louis, MO, USA, 1989; pp. 895–898. [Google Scholar]
- Kaplan, L.A. Glucose. In Clinical Chemistry: Theory, Analysis, and Correlation; Kaplan, L.A., Pesce, A.J., Eds.; The CV Mosby Company: St. Louis, MO, USA, 1989; p. 436. [Google Scholar]
- Schultz, A. Uric acid. In Clinical Chemistry: Theory, Analysis, and Correlation, 2nd ed.; Kaplan, L.A., Pesce, A.J., Eds.; The CV Mosby Company: St. Louis, MO, USA, 1989; pp. 1261–1266. [Google Scholar]
- Kaplan, L.A. Urea. In Clinical Chemistry: Theory, Analysis, and Correlation, 2nd ed.; Kaplan, L.A., Pesce, A.J., Eds.; The CV Mosby Company: St. Louis, MO, USA, 1989; pp. 1257–1260. [Google Scholar]
- DeChatelet, L.R.; McCall, C.E.; McPhail, L.C.; Johnston, R.B. Superoxide Dismutase Activity in Leukocytes. J. Clin. Investig. 1974, 53, 1197–1201. [Google Scholar] [CrossRef]
- Beutler, E.; Duron, O.; Kelly, B.M. Improved method for the determination of blood glutathione. J. Lab. Clin. Med. 1963, 61, 882–888. [Google Scholar]
- Aebi, H. [13] Catalase in vitro. In Methods in Enzymology; Elsevier Inc.: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Koracevic, D.; Harris, G.; Rayner, A.; Blair, J.; Watt, B.; Koracevic, G.; Djordjevic, V.; Andrejevic, S.; Cosic, V. Method for the measurement of antioxidant activity in human fluids. J. Clin. Pathol. 2001, 54, 356–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badawy, A.A.; El-Magd, M.A.; Al Sadrah, S.A. Therapeutic Effect of Camel Milk and Its Exosomes on MCF7 Cells In Vitro and In Vivo. Integr. Cancer Ther. 2018, 17, 1235–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resnier, P.; Galopin, N.; Sibiril, Y.; Clavreul, A.; Cayon, J.; Briganti, A.; Legras, P.; Vessieres, A.; Montier, T.; Jaouen, G.; et al. Efficient ferrocifen anticancer drug and Bcl-2 gene therapy using lipid nanocapsules on human melanoma xenograft in mouse. Pharmacol. Res. 2017, 126, 54–65. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of compounds (1) and (2) are available from the authors. |









| Complex | Kq (M−1) | Kapp (M−1) |
|---|---|---|
| (1) | 1.96 ± 0.04 × 103 | 1.08 ± 0.11 × 107 |
| (2) | 8.64 ± 0.15 × 103 | 1.72 ± 0.28 × 107 |
| Compound | MCF-7 | Caco2 | THLE-2 | |||
|---|---|---|---|---|---|---|
| µg/mL | µM | µg/mL | µM | µg/mL | µM | |
| Hapbim | 61 ± 4 | 290 ± 20 | 80 ± 5 | 380 ± 20 | 1140 ± 80 | 5500 ± 100 |
| (1) | 170 ± 10 | 320 ± 20 | 155 ± 9 | 290 ± 20 | 990 ± 70 | 1800 ± 100 |
| (2) | 118 ± 8 | 230 ± 10 | 129 ± 8 | 250 ± 10 | 1280 ± 80 | 2500 ± 100 |
| Cisplatin | 22 ± 1 | 73 ± 5 | 18 ± 1 | 60 ± 5 | 650 ± 60 | 2200 ± 100 |
| Therapeutic Index | Hapbim | (1) | (2) | Cisplatin |
|---|---|---|---|---|
| IC50 of THLE-2 cell/IC50 of MCF-7 | 18.5 | 5.8 | 10.89 | 29.3 |
| IC50 of THLE-2 cell/IC50 ofCaco2 | 14.2 | 6.4 | 9.95 | 35.9 |
| Group | GOT (U/L) | GPT (U/L) | Albumin (g/dL) | Bilirubin (mg/dL) | Creatinine (mg/dL) | Urea (mg/dL) | Uric Acid (mg/dL) |
|---|---|---|---|---|---|---|---|
| Control | 36.1 ± 2.4 | 16.7 ± 0.6 | 2.9 ± 0.03 | 0.13 ± 0.02 | 0.4 ± 0.03 | 35.4 ± 0.5 | 2.85 ± 0.04 |
| I control | 41.8 ± 2.9 b | 15.6 ± 1.6 b | 2.3 ± 0.1 * | 0.15 ±0.02 b | 0.37 ±0.1 b | 33.5 ± 0.7 b | 2.9 ± 0.1 b |
| II control | 54.9 ± 2.2 * | 17.2 ± 0.5 b | 2.7 ± 0.1 b | 0.13 ± 0.02 b | 0.7 ± 0.02 * | 31.7 ± 1.2 b | 2.97 ± 0.1 b |
| III control | 68.8 ± 3.6 * | 19.6 ± 0.3 * | 2.3 ± 0.1 * | 0.26 ±0.02 * | 0.75±0.02 * | 36.7 ± 1.6 b | 2.77 ± 0.1 b |
| EAC | 250 ± 42.1 * | 93.7 ± 4.8 * | 0.5 ± 0.1 * | 0.3 ± 0.03 * | 1.1 ± 0.1 * | 41.5 ±0.5 * | 3.74 ± 0.04 * |
| I treated | 248 ± 15.6 a | 15.2 ± 0.7 # | 0.9 ± 0.03 # | 0.18 ± 0.03 # | 0.7 ± 0.01 # | 34.5 ± 0.4 # | 2.6 ± 0.1 # |
| II treated | 179 ± 10.8 # | 17.8 ± 0.6 # | 2.7 ± 0.3 # | 0.2 ± 0.03 # | 0.7 ± 0.1 # | 34.2 ± 0.6 # | 2.7 ± 0.1 # |
| III treated | 145 ± 39.3 # | 31.9 ± 7.8 # | 2.6 ± 0.1 # | 0.18 ±0.02 # | 1 ± 0.1 a | 33.3 ± 1.2 # | 2.8 ± 0.1 # |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Bax | ACACCTGAGCTGACCTTG | AGCCCATGATGGTTCTGATC |
| Bcl2 | AGTACCTGAACCGGCATCTG | CATGCTGGGGCCATATAGTT |
| Caspase 3 | TTAATAAAGGTATCCATGGAGAACACT | TTAGTGATAAAAATAGAGTTCTTTTGTGAG |
| β actin | AAGTCCCTCACCCTCCCAAAAG | AAGCAATGCTGTCACCTTCCC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elsayed, S.A.; Harrypersad, S.; Sahyon, H.A.; El-Magd, M.A.; Walsby, C.J. Ruthenium(II)/(III) DMSO-Based Complexes of 2-Aminophenyl Benzimidazole with In Vitro and In Vivo Anticancer Activity. Molecules 2020, 25, 4284. https://doi.org/10.3390/molecules25184284
Elsayed SA, Harrypersad S, Sahyon HA, El-Magd MA, Walsby CJ. Ruthenium(II)/(III) DMSO-Based Complexes of 2-Aminophenyl Benzimidazole with In Vitro and In Vivo Anticancer Activity. Molecules. 2020; 25(18):4284. https://doi.org/10.3390/molecules25184284
Chicago/Turabian StyleElsayed, Shadia A., Shane Harrypersad, Heba A. Sahyon, Mohammed Abu El-Magd, and Charles J. Walsby. 2020. "Ruthenium(II)/(III) DMSO-Based Complexes of 2-Aminophenyl Benzimidazole with In Vitro and In Vivo Anticancer Activity" Molecules 25, no. 18: 4284. https://doi.org/10.3390/molecules25184284