Feature Papers in Biomacromolecules: Nucleic Acids
Share This Topical Collection
Editor
 Prof. Dr. Peter E. Nielsen
Prof. Dr. Peter E. Nielsen
 Prof. Dr. Peter E. Nielsen
Prof. Dr. Peter E. Nielsen
E-Mail
Website
Collection Editor
Department of Cellular and Molecular Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Blegdamsvej 3C, DK-2200 Copenhagen, Denmark
Interests: antisense; drug delivery; antibiotics; peptide nucleic acid; gene targeting
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
This Topical Collection, “Feature Papers in Biomacromolecules: Nucleic Acids”, will bring together high-quality research articles, review articles, and communications on all aspects of nucleic acids. It is dedicated to diverse recent advances in nucleic acids research, and comprises a selection of exclusive papers from the Editorial Board Members (EBMs) of the Biomacromolecules: Nucleic Acids Section as well as invited papers from relevant experts. We also welcome established experts in the field to make contributions to this Topical Collection. Please note that all invited papers will be published online once accepted. We aim to represent our Section as an attractive open access publishing platform for nucleic acids research.
Prof. Dr. Peter E. Nielsen
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Biomolecules is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Published Papers (4 papers)
Open AccessCommunication
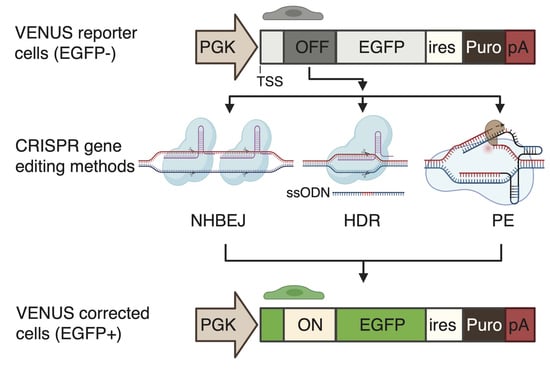
Comparison of In-Frame Deletion, Homology-Directed Repair, and Prime Editing-Based Correction of Duchenne Muscular Dystrophy Mutations
by
Xiaoying Zhao, Kunli Qu, Benedetta Curci, Huanming Yang, Lars Bolund, Lin Lin and Yonglun Luo
Cited by 1 | Viewed by 2348
Abstract
Recent progress in CRISPR gene editing tools has substantially increased the opportunities for curing devastating genetic diseases. Here we compare in-frame deletion by CRISPR-based non-homologous blunt end joining (NHBEJ), homology-directed repair (HDR), and prime editing (PE, PE2, and PE3)-based correction of two Duchenne
[...] Read more.
Recent progress in CRISPR gene editing tools has substantially increased the opportunities for curing devastating genetic diseases. Here we compare in-frame deletion by CRISPR-based non-homologous blunt end joining (NHBEJ), homology-directed repair (HDR), and prime editing (PE, PE2, and PE3)-based correction of two Duchenne Muscular Dystrophy (DMD) loss-of-function mutations (c.5533G>T and c.7893delC). To enable accurate and rapid evaluation of editing efficiency, we generated a genomically integrated synthetic reporter system (VENUS) carrying the
DMD mutations. The VENUS contains a modified enhanced green fluorescence protein (
EGFP) gene, in which expression was restored upon the CRISPR-mediated correction of
DMD loss-of-function mutations. We observed that the highest editing efficiency was achieved by NHBEJ (74–77%), followed by HDR (21–24%) and PE2 (1.5%) in HEK293T VENUS reporter cells. A similar HDR (23%) and PE2 (1.1%) correction efficiency is achieved in fibroblast VENUS cells. With PE3 (PE2 plus nicking gRNA), the c.7893delC correction efficiency was increased 3-fold. Furthermore, an approximately 31% correction efficiency of the endogenous DMD: c.7893delC is achieved in the FACS-enriched HDR-edited VENUS EGFP+ patient fibroblasts. We demonstrated that a highly efficient correction of
DMD loss-of-function mutations in patient cells can be achieved by several means of CRISPR gene editing.
Full article
►▼
Show Figures
Open AccessReview
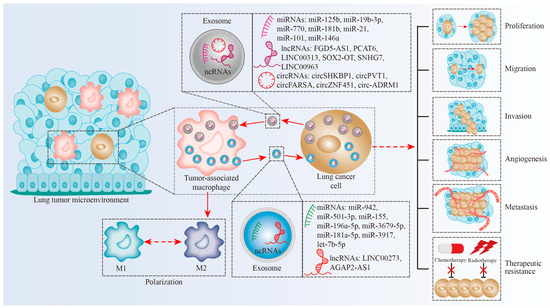
Exosomal Non-Coding RNAs: Novel Regulators of Macrophage-Linked Intercellular Communication in Lung Cancer and Inflammatory Lung Diseases
by
Xingning Lai, Jie Zhong, Boyi Zhang, Tao Zhu and Ren Liao
Cited by 3 | Viewed by 2317
Abstract
Macrophages are innate immune cells and often classified as M1 macrophages (pro-inflammatory states) and M2 macrophages (anti-inflammatory states). Exosomes are cell-derived nanovesicles that range in diameter from 30 to 150 nm. Non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular
[...] Read more.
Macrophages are innate immune cells and often classified as M1 macrophages (pro-inflammatory states) and M2 macrophages (anti-inflammatory states). Exosomes are cell-derived nanovesicles that range in diameter from 30 to 150 nm. Non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs), are abundant in exosomes and exosomal ncRNAs influence immune responses. Exosomal ncRNAs control macrophage-linked intercellular communication via their targets or signaling pathways, which can play positive or negative roles in lung cancer and inflammatory lung disorders, including acute lung injury (ALI), asthma, and pulmonary fibrosis. In lung cancer, exosomal ncRNAs mediated intercellular communication between lung tumor cells and tumor-associated macrophages (TAMs), coordinating cancer proliferation, migration, invasion, metastasis, immune evasion, and therapy resistance. In inflammatory lung illnesses, exosomal ncRNAs mediate macrophage activation and inflammation to promote or inhibit lung damage. Furthermore, we also discussed the possible applications of exosomal ncRNA-based therapies for lung disorders.
Full article
►▼
Show Figures
Open AccessReview
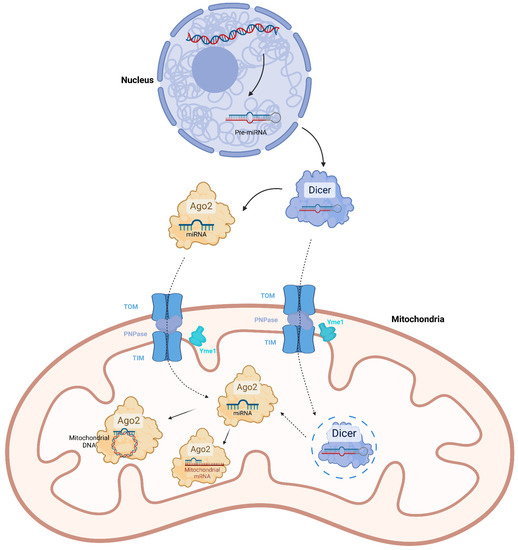
Mitochondrial Non-Coding RNAs Are Potential Mediators of Mitochondrial Homeostasis
by
Weihan Sun, Yijian Lu, Heng Zhang, Jun Zhang, Xinyu Fang, Jianxun Wang and Mengyang Li
Cited by 7 | Viewed by 2348
Abstract
Mitochondria are the energy production center in cells, which regulate aerobic metabolism, calcium balance, gene expression and cell death. Their homeostasis is crucial for cell viability. Although mitochondria own a nucleus-independent and self-replicating genome, most of the proteins, which fulfill mitochondrial functions and
[...] Read more.
Mitochondria are the energy production center in cells, which regulate aerobic metabolism, calcium balance, gene expression and cell death. Their homeostasis is crucial for cell viability. Although mitochondria own a nucleus-independent and self-replicating genome, most of the proteins, which fulfill mitochondrial functions and mitochondrial quality control, are encoded by the nuclear genome and are imported into mitochondria. Hence, the regulation of mitochondrial protein expression and translocation is considered essential for mitochondrial homeostasis. By means of high-throughput RNA sequencing and bioinformatic analysis, non-coding RNAs localized in mitochondria have been generally identified. They are either generated from the mitochondrial genome or the nuclear genome. The mitochondrial non-coding RNAs can directly interact with mitochondrial DNAs or transcripts to affect gene expression. They can also bind nuclear genome-encoded mitochondrial proteins to regulate their mitochondrial import, protein level and combination. Generally, mitochondrial non-coding RNAs act as regulators for mitochondrial processes including oxidative phosphorylation and metabolism. In this review, we would like to introduce the latest research progressions regarding mitochondrial non-coding RNAs and summarize their identification, biogenesis, translocation, molecular mechanism and function.
Full article
►▼
Show Figures
Open AccessEditor’s ChoiceReview
Synthesis of Protein-Oligonucleotide Conjugates
by
Emma E. Watson and Nicolas Winssinger
Cited by 2 | Viewed by 3003
Abstract
Nucleic acids and proteins form two of the key classes of functional biomolecules. Through the ability to access specific protein-oligonucleotide conjugates, a broader range of functional molecules becomes accessible which leverages both the programmability and recognition potential of nucleic acids and the structural,
[...] Read more.
Nucleic acids and proteins form two of the key classes of functional biomolecules. Through the ability to access specific protein-oligonucleotide conjugates, a broader range of functional molecules becomes accessible which leverages both the programmability and recognition potential of nucleic acids and the structural, chemical and functional diversity of proteins. Herein, we summarize the available conjugation strategies to access such chimeric molecules and highlight some key case study examples within the field to showcase the power and utility of such technology.
Full article
►▼
Show Figures