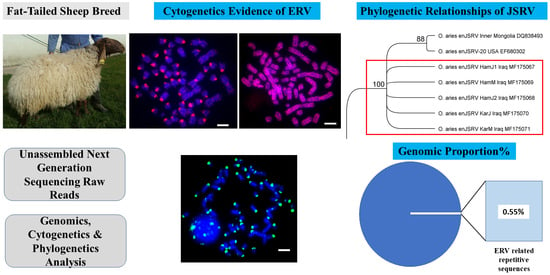
The Nature and Chromosomal Landscape of Endogenous Retroviruses (ERVs) Integrated in the Sheep Nuclear Genome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Materials
2.2. Discovery of Endogenous Retrovirus (ERV)-Related Repetitive Sequences
2.2.1. Graph-Based Read Clustering (RepeatExplorer)
2.2.2. k-mer Frequency Tool (Jellyfish)
2.2.3. Database Searching
2.3. Design and Amplification of FISH Probes, Chromosome Preparation and In Situ Hybridization
2.4. Assembly of the Complete Genome of the Endogenous Jaagsiekte Sheep Retrovirus (enJSRV)
2.5. Data Analysis and Phylogenetic Relationships
3. Results
3.1. Identification of Endogenous Retroviruses Related Sequences
3.2. Identification and Quantification of ERV Repeats in Ancestral and Bos Taurus ERV Sequences
3.3. Abundance and Genomic Organization ERV-Related Repetitive Elements
3.3.1. ERV1
3.3.2. ERV2
3.3.3. ERV3
3.3.4. Combined ERV1 and Satellite like Sequences
3.4. The Complete Genome of the Endogenous Jaagsiekte Sheep Retrovirus (enJSRV)
4. Discussion
4.1. Genomic Distribution and Chromosomal Organization of ERVs
4.2. Complete Genome of Endogenous Betaretroviruses (enJSRV) and Their Abundance in Iraqi Sheep Breeds
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. (Pat) Repetitive DNA in Eukaryotic Genomes. Chromosom. Res. 2015, 23, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; Fitzhugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikkelsen, T.S.; Hillier, L.W.; Eichler, E.E.; Zody, M.C.; Jaffe, D.B.; Yang, S.P.; Enard, W.; Hellmann, I.; Lindblad-Toh, K.; Altheide, T.K.; et al. Initial Sequence of the Chimpanzee Genome and Comparison with the Human Genome. Nature 2005, 437, 69–87. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A Unified Classification System for Eukaryotic Transposable Elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Mager, D.L.; Stoye, J.P. Mammalian Endogenous Retroviruses. Mob. DNA III 2015, 1079–1100. [Google Scholar] [CrossRef]
- Feschotte, C.; Gilbert, C. Endogenous Viruses: Insights into Viral Evolution and Impact on Host Biology. Nat. Rev. Genet. 2012, 13, 283–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannert, N.; Kurth, R. The Evolutionary Dynamics of Human Endogenous Retroviral Families. Annu. Rev. Genomics Hum. Genet. 2006, 7, 149–173. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.A. The Discovery of Endogenous Retroviruses. Retrovirology 2006, 3, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Kojima, K.K. Human Transposable Elements in Repbase: Genomic Footprints from Fish to Humans. Mob. DNA 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuo, X.; Feschotte, C. Cross-Species Transmission and Differential Fate of an Endogenous Retrovirus in Three Mammal Lineages. PLoS Pathog. 2015, 11, e1005279. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Montojo, M.; Doucet-O’Hare, T.; Henderson, L.; Nath, A. Human Endogenous Retrovirus-K (HML-2): A Comprehensive Review. Crit. Rev. Microbiol. 2018, 44, 715–738. [Google Scholar] [CrossRef] [PubMed]
- Katoh, I.; Kurata, S.I. Association of Endogenous Retroviruses and Long Terminal Repeats with Human Disorders. Front. Oncol. 2013, 3, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balestrieri, E.; Pica, F.; Matteucci, C.; Zenobi, R.; Sorrentino, R.; Argaw-Denboba, A.; Cipriani, C.; Bucci, I.; Sinibaldi-Vallebona, P. Transcriptional Activity of Human Endogenous Retroviruses in Human Peripheral Blood Mononuclear Cells. Biomed Res. Int. 2015, 2015, 164529. [Google Scholar] [CrossRef] [PubMed]
- Grandi, N.; Tramontano, E. Type W Human Endogenous Retrovirus (HERV-W) Integrations and Their Mobilization by L1 Machinery: Contribution to the Human Transcriptome and Impact on the Host Physiopathology. Viruses 2017, 9, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnaud, F.; Caporale, M.; Varela, M.; Biek, R.; Chessa, B.; Alberti, A.; Golder, M.; Mura, M.; Zhang, Y.P.; Yu, L.; et al. A Paradigm for Virus-Host Coevolution: Sequential Counter-Adaptations between Endogenous and Exogenous Retroviruses. PLoS Pathog. 2007, 3, 1716–1729. [Google Scholar] [CrossRef] [PubMed]
- Palmarini, M.; Mura, M.; Spencer, T.E. Endogenous Betaretroviruses of Sheep: Teaching New Lessons in Retroviral Interference and Adaptation. J. Gen. Virol. 2004, 85, 1–13. [Google Scholar] [CrossRef]
- Murcia, P.R.; Arnaud, F.; Palmarini, M. The Transdominant Endogenous Retrovirus EnJS56A1 Associates with and Blocks Intracellular Trafficking of Jaagsiekte Sheep Retrovirus Gag. J. Virol. 2007, 81, 1762–1772. [Google Scholar] [CrossRef] [Green Version]
- Qi, J.W.; Xu, M.J.; Liu, S.Y.; Zhang, Y.F.; Liu, Y.; Zhang, Y.K.; Cao, G.F. Identification of Sheep Endogenous Beta-Retroviruses with Uterus-Specific Expression in the Pregnant Mongolian Ewe. J. Integr. Agric. 2013, 12, 884–891. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Macas, J. Graph-Based Clustering and Characterization of Repetitive Sequences in next-Generation Sequencing Data. BMC Bioinformatics 2010, 11, 378. [Google Scholar] [CrossRef] [Green Version]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-Based Web Server for Genome-Wide Characterization of Eukaryotic Repetitive Elements from next-Generation Sequence Reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marçais, G.; Kingsford, C. A Fast, Lock-Free Approach for Efficient Parallel Counting of Occurrences of k-Mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, S.Y.; Li, J.Y.; Han, M.; Wang, Z.L. Cloning and Sequence Analysis of Genome from the Inner Mongolia Strain of the Endogenous Betaretroviruses (EnJSRV). Virol. Sin. 2008, 23, 15–24. [Google Scholar] [CrossRef]
- Contreras-Galindo, R.; Kaplan, M.H.; He, S.; Contreras-Galindo, A.C.; Gonzalez-Hernandez, M.J.; Kappes, F.; Dube, D.; Chan, S.M.; Robinson, D.; Meng, F.; et al. HIV Infection Reveals Widespread Expansion of Novel Centromeric Human Endogenous Retroviruses. Genome Res. 2013, 23, 1505–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zahn, J.; Kaplan, M.H.; Fischer, S.; Dai, M.; Meng, F.; Saha, A.K.; Cervantes, P.; Chan, S.M.; Dube, D.; Omenn, G.S.; et al. Expansion of a Novel Endogenous Retrovirus throughout the Pericentromeres of Modern Humans. Genome Biol. 2015, 16, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prudhomme, S.; Bonnaud, B.; Mallet, F. Endogenous Retroviruses and Animal Reproduction. Cytogenet. Genome Res. 2005, 110, 353–364. [Google Scholar] [CrossRef]
- Chaves, R.; Guedes-Pinto, H.; Heslop-Harrison, J.S.; Schwarzacher, T. The Species and Chromosomal Distribution of the Centromeric α-Satellite I Sequence from Sheep in the Tribe Caprini and Other Bovidae. Cytogenet. Genome Res. 2000, 91, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Ferreri, G.C.; Brown, J.D.; Obergfell, C.; Jue, N.; Finn, C.E.; O’Neill, M.J.; O’Neill, R.J. Recent Amplification of the Kangaroo Endogenous Retrovirus, KERV, Limited to the Centromere. J. Virol. 2011, 85, 4761–4771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lomiento, M.; Jiang, Z.; D’Addabbo, P.; Eichler, E.E.; Rocchi, M. Evolutionary-New Centromeres Preferentially Emerge within Gene Deserts. Genome Biol. 2008, 9, R173. [Google Scholar] [CrossRef] [Green Version]
- Escudeiro, A.; Ferreira, D.; Mendes-da-Silva, A.; Heslop-Harrison, J.S.; Adega, F.; Chaves, R. Bovine Satellite DNAs–a History of the Evolution of Complexity and Its Impact in the Bovidae Family. Eur. Zool. J. 2019, 86, 20–37. [Google Scholar] [CrossRef]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a Database of Eukaryotic Repetitive Elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a Database of Repetitive Elements in Eukaryotic Genomes. Mob. DNA 2015, 6, 4–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Etxebarria, K.; Jugo, B.M. Evolutionary History of Bovine Endogenous Retroviruses in the Bovidae Family. BMC Evol. Biol. 2013, 13, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pačes, J.; Pavlícacek, A.; Pačes, V. HERVd: Database of Human Endogenous Retroviruses. Nucleic Acids Res. 2002, 30, 205–206. [Google Scholar] [CrossRef] [Green Version]
- Jern, P.; Coffin, J.M. Effects of Retroviruses on Host Genome Function. Annu. Rev. Genet. 2008, 42, 709–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varela, M.; Spencer, T.E.; Palmarini, M.; Arnaud, F. Friendly Viruses: The Special Relationship between Endogenous Retroviruses and Their Host. Ann. N. Y. Acad. Sci. 2009, 1178, 157–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurth, R.; Bannert, N. Beneficial and Detrimental Effects of Human Endogenous Retroviruses. Int. J. Cancer 2010, 126, 306–314. [Google Scholar] [CrossRef]
- Harper, G.; Osuji, J.O.; Heslop-Harrison, J.S.; Hull, R. Integration of Banana Streak Badnavirus into the Musa Genome: Molecular and Cytogenetic Evidence. Virology 1999, 255, 207–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noreen, F.; Akbergenov, R.; Hohn, T.; Richert-Pöggeler, K.R. Distinct Expression of Endogenous Petunia Vein Clearing Virus and the DNA Transposon DTph1 in Two Petunia Hybrida Lines Is Correlated with Differences in Histone Modification and SiRNA Production. Plant J. 2007, 50, 219–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cumer, T.; Pompanon, F.; Boyer, F. Old Origin of a Protective Endogenous Retrovirus (EnJSRV) in the Ovis Genus. Heredity 2019, 122, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, K.A.; Palmarini, M.; Adelson, D.L.; Spencer, T.E. Sheep Endogenous Betaretroviruses (EnJSRVs) and the Hyaluronidase 2 (HYAL2) Receptor in the Ovine Uterus and Conceptus. Biol. Reprod. 2005, 73, 271–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunlap, K.A.; Palmarini, M.; Spencer, T.E. Ovine Endogenous Betaretroviruses (EnJSRVs) and Placental Morphogenesis. Placenta 2006, 27, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Etxebarria, K.; Jugo, B.M. Genome-Wide Detection and Characterization of Endogenous Retroviruses in Bos Taurus. J. Virol. 2010, 84, 10852–10862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sistiaga-Poveda, M.; Jugo, B.M. Evolutionary Dynamics of Endogenous Jaagsiekte Sheep Retroviruses Proliferation in the Domestic Sheep, Mouflon and Pyrenean Chamois. Heredity (Edinb). 2014, 112, 571–578. [Google Scholar] [CrossRef]
- Nikolai, K.; Mathias, M.; Gottfried, B.; Bernhard, A. Characterization of Endogenous Retroviruses in Sheep. J. Virol. 2003, 77, 11268–11273. [Google Scholar] [CrossRef] [Green Version]






| Probe Names | Name of Primers [Sequence (5’–3’)] | Expected Product Size (bp) | Annealing Temp. |
|---|---|---|---|
| CL18C5_ERV1 | F = ATCTTGGCTGAGCGATGCG | 246 | 62 |
| R = GGGCTCTTGTCTAACACTCGG | |||
| CL20C5_ERV1 | F = TGTGTTGCCATGACCACTCC | 574 | 62 |
| R = TGCCAGCATTCTTGGACTCC | |||
| CL23C4_ERV1 | F = CAAGGAATTTGGAGTGGTGGG | 195 | 62 |
| R = TCGGTGGTCCTGTTGTAGCC | |||
| CL25_ERV1 | F = TGTCATCTGGTCACTGCTGC | 402 | 62 |
| R = AGGGAGTTTGCAGGATGTGG | |||
| 22mer_ERV1.A | F = CACTCTTTTGCCCAATCCGG | 545 | 60 |
| R = CAGCTACTTTTCGAGCTGCC | |||
| 32mer_ERV1.RE | F = GGTTTTAGATGGGACCGGGC | 564 | 62 |
| R = TCTTCCTGCCATTCGAAGGC | |||
| 32mer_ERV1.T3 | F = TGCTTCTTTTCAACGCACCC | 541 | 64 |
| R = CTTGATGGAGCCAGGTACCC | |||
| CL14C75_ERV2 | F = GGTGATTTACATCATCTTCTGGCC | 505 | 62 |
| R = AGCTTGCCTAACAGGTTCCC | |||
| OuttopCL_ERV2 | F = AAAGGTCACGAGGATGAGGC | 555 | 60 |
| R = AGGACAAAGGTGCAGTGGG | |||
| CL37_ERV2 | F = TGTCTTTTCCTCTCCTCGGC | 488 | 62 |
| R = CATGCTTATGTCTGGGCTGC | |||
| CL67_ERV3 | F = ATTCAATCTCCTAATATTCCCACCC | 315 | 58 |
| R = GTTAGTAGTCAAGCTTTTGTCTGGC | |||
| CL27C1_ERV1+ERV3 | F = GCAGGTCGGTGTATCTTCCC | 619 | 62 |
| R = GGGAACTTGCAAGAGTGGGG | |||
| 32mer_ERV1+ERV3 | F = CTTGCAAGAGTGGGGAAAGC | 615 | 62 |
| R = GCAGGTCGGTGTATCTTCCC | |||
| 32mer_ERV1+CRC | F = TACAGAGCAAAGGGGATGGG | 468 | 60 |
| R = TGGTTGTTTCTTTCCACCATTCC |
| Probes of Endogenous Retroviruses ERVs | PCR Product bp | HamJ1_Male Genome | KarJ_Female Genome | ||||
|---|---|---|---|---|---|---|---|
| Assembled Reads | Copies of Probe | Genomic Proportion % | Assembled Reads | Copies of Probe | Genomic Proportion % | ||
| CL18C5_ERV1 | 246 | 5973 | 3642 | 0.0115 | 7525 | 4588 | 0.0124 |
| CL20C5_ERV1 | 574 | 3124 | 816 | 0.0060 | 3676 | 961 | 0.0061 |
| CL23C4_ERV1 | 195 | 2157 | 1659 | 0.0041 | 2000 | 1538 | 0.0033 |
| CL25_ERV1 | 402 | 6900 | 2575 | 0.0133 | 4364 | 1628 | 0.0072 |
| 22mer_ERV1.A | 545 | 3452 | 950 | 0.0066 | 3008 | 828 | 0.0050 |
| 32mer_ERV1.RE | 564 | 3753 | 998 | 0.0072 | 3426 | 911 | 0.0057 |
| 32mer_ERV1.T3 | 541 | 1709 | 474 | 0.0033 | 2554 | 708 | 0.0042 |
| CL14C75_ERV2 | 505 | 5294 | 1572 | 0.0102 | 6943 | 2062 | 0.0115 |
| OuttopCL_ERV2 | 555 | 1299 | 351 | 0.0025 | 1177 | 318 | 0.0019 |
| CL37_ERV2 | 488 | 2113 | 649 | 0.0041 | 2338 | 719 | 0.0039 |
| CL67_ERV3 | 315 | 1000 | 476 | 0.0019 | 0 | 0 | 0 |
| CL27C1_ERV1+ERV3 | 619 | 5907 | 1431 | 0.0113 | 5595 | 1356 | 0.0092 |
| 32mer_ERV1+ERV3 | 615 | 5500 | 1341 | 0.0106 | 5300 | 1293 | 0.0087 |
| 32mer_ERV1+CRC | 468 | 5700 | 1827 | 0.0110 | 5760 | 1846 | 0.0095 |
| Clusters | Total Length | Number of Reads | Genome Proportion [%] | Repbase Database Similarities | Graph Layout |
|---|---|---|---|---|---|
| CL14C75_ERV2 | 309,579 | 2057 | 0.229 | ERV2 |  |
| CL18C5_ERV1 | 32,673 | 217 | 0.024 | ERV1 |  |
| CL20C5_ERV1 | 28,460 | 189 | 0.021 | ERV1 |  |
| CL23C4_ERV1 | 21,077 | 140 | 0.016 | ERV1 |  |
| CL27C1_ERV3+ERV1 | 17,746 | 118 | 0.013 | ERV3 and ERV1 |  |
| CL25_ERV1 | 18,501 | 123 | 0.0147 | ERV1 |  |
| CL37_ERV2 | 9926 | 66 | 0.0079 | ERV2 |  |
| ERV Probes | Chromosomes | Localization | Figures |
|---|---|---|---|
| CL18C5_ERV1 | All chromosomes except XY and the largest pair of submetacentrics | Centromere | 1 |
| CL20C5_ERV1 | All chromosomes except XY and the largest pair of submetacentrics | Centromere | |
| CL23C4_ERV1 | All chromosomes | Centromere to disperse | |
| CL25_ERV1 | All chromosomes | Centromere- and dispersed-like dots | |
| 22mer_ERV1.A | All chromosomes | Centromere of all acrocentrics, XY, pair of submetacentrics; Telomere of other 2 pairs of submetacentric | 2 |
| 32mer_ERV1.RE | All chromosomes | Centromere to disperse | |
| 32mer_ERV1.T3 | All chromosomes | Centromere to disperse | |
| CL14C75_ERV2 | All chromosomes | Dispersed | 3 |
| OuttopCL_ERV2 | All chromosomes | Centromere to disperse | |
| CL37_ERV2 | All chromosomes | Centromere to disperse | |
| CL67_ERV3 | All chromosomes | Centromere- and dispersed-like dots | 4 |
| CL27C1_ERV1+ERV3 | All chromosomes except XY | Centromere and subtelomere | 5 |
| 32mer_ERV1+ERV3 | All chromosomes | Centromere to disperse and few telomere | |
| 32mer_ERV1+CRC | Most chromosomes | Centromere and few telomere |
| enJSRV Breeds (GenBank ID) | Complete enJSRV Genome bp | Assembled Reads | Total Reads of Each Genome (Coverage X) | Genomic Proportion % | Copies of enJSRV (Assembled Reads*150/7941) |
|---|---|---|---|---|---|
| HamJ1 (MF175067) | 7941 | 5390 | 52,048,068 (2.6) | 0.0104 | 101.81 |
| HamJ2 (MF175068) | 7941 | 6606 | 56,220,882 (2.81) | 0.0118 | 124.78 |
| HamM (MF175069) | 7941 | 3809 | 43,596,654 (2.18) | 0.0087 | 71.95 |
| KaJ (MF175070) | 7941 | 5386 | 60,605,648 (3.03) | 0.0089 | 101.74 |
| KarM (MF175071) | 7941 | 4846 | 44,933,034 (2.25) | 0.0108 | 91.54 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mustafa, S.I.; Schwarzacher, T.; Heslop-Harrison, J.S. The Nature and Chromosomal Landscape of Endogenous Retroviruses (ERVs) Integrated in the Sheep Nuclear Genome. DNA 2022, 2, 86-103. https://doi.org/10.3390/dna2010007
Mustafa SI, Schwarzacher T, Heslop-Harrison JS. The Nature and Chromosomal Landscape of Endogenous Retroviruses (ERVs) Integrated in the Sheep Nuclear Genome. DNA. 2022; 2(1):86-103. https://doi.org/10.3390/dna2010007
Chicago/Turabian StyleMustafa, Sarbast Ihsan, Trude Schwarzacher, and John S. Heslop-Harrison. 2022. "The Nature and Chromosomal Landscape of Endogenous Retroviruses (ERVs) Integrated in the Sheep Nuclear Genome" DNA 2, no. 1: 86-103. https://doi.org/10.3390/dna2010007