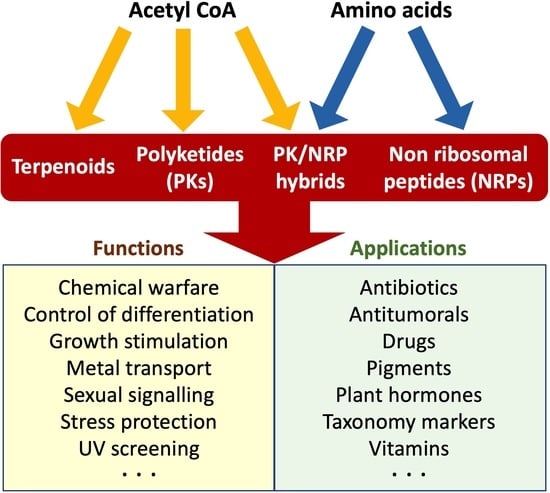
Fungal Secondary Metabolism
Definition
:1. Introduction
2. Chemical Families of SMs
2.1. Polyketides
2.2. Non-Ribosomal Peptides
2.3. Hybrid Non-Ribosomal Peptide/Polyketides
2.4. Terpenoids
3. Genetic Organization and Regulation of SM Genes
4. Biological Functions
5. Biological Properties and Applications
6. Conclusions and Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Entry Link on the Encyclopedia Platform
References
- Raistrick, H. A Region of Biosynthesis. Proc. R. Soc. Lond. B Biol. Sci. 1950, 136, 481–508. [Google Scholar] [CrossRef]
- Bennett, J.W.; Chung, K.T. Alexander Fleming and the Discovery of Penicillin. Adv. Appl. Microbiol. 2001, 49, 163–184. [Google Scholar] [CrossRef]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal Secondary Metabolism - from Biochemistry to Genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef]
- Robey, M.T.; Caesar, L.K.; Drott, M.T.; Keller, N.P.; Kelleher, N.L. An Interpreted Atlas of Biosynthetic Gene Clusters from 1,000 Fungal Genomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2020230118. [Google Scholar] [CrossRef] [PubMed]
- Staunton, J.; Weissman, K.J. Polyketide Biosynthesis: A Millennium Review. Nat. Prod. Rep. 2001, 18, 380–416. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, B.J. Biosynthesis of Polyketides. Nat. Prod. Rep. 1997, 14, 523–556. [Google Scholar] [CrossRef] [PubMed]
- Shen, B. Polyketide Biosynthesis beyond the Type I, II and III Polyketide Synthase Paradigms. Curr. Opin. Chem. Biol. 2003, 7, 285–295. [Google Scholar] [CrossRef]
- Herbst, D.A.; Townsend, C.A.; Maier, T. The Architectures of Iterative Type I PKS and FAS. Nat. Prod. Rep. 2018, 35, 1046–1069. [Google Scholar] [CrossRef]
- Weissman, K.J. Uncovering the Structures of Modular Polyketide Synthases. Nat. Prod. Rep. 2015, 32, 436–453. [Google Scholar] [CrossRef]
- Kim, Y.-T.; Lee, Y.-R.; Jin, J.; Han, K.-H.; Kim, H.; Kim, J.-C.; Lee, T.; Yun, S.-H.; Lee, Y.-W. Two Different Polyketide Synthase Genes Are Required for Synthesis of Zearalenone in Gibberella zeae. Mol. Microbiol. 2005, 58, 1102–1113. [Google Scholar] [CrossRef]
- Nelson, P.E.; Desjardins, A.E.; Plattner, R.D. Fumonisins, Mycotoxins Produced by Fusarium Species: Biology, Chemistry, and Significance. Annu. Rev. Phytopathol. 1993, 31, 233–252. [Google Scholar] [CrossRef] [PubMed]
- Mulder, K.C.L.; Mulinari, F.; Franco, O.L.; Soares, M.S.F.; Magalhães, B.S.; Parachin, N.S. Lovastatin Production: From Molecular Basis to Industrial Process Optimization. Biotechnol. Adv. 2015, 33, 648–665. [Google Scholar] [CrossRef]
- Klich, M.A. Aspergillus flavus: The Major Producer of Aflatoxin. Mol. Plant Pathol. 2007, 8, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Limón, M.C.; Rodríguez-Ortiz, R.; Avalos, J. Bikaverin Production and Applications. Appl. Microbiol. Biotechnol. 2010, 87, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Prosperini, A.; Berrada, H.; Ruiz, M.J.; Caloni, F.; Coccini, T.; Spicer, L.J.; Perego, M.C.; Lafranconi, A. A Review of the Mycotoxin Enniatin B. Front. Public Health 2017, 5, 304. [Google Scholar] [CrossRef]
- Survase, S.A.; Kagliwal, L.D.; Annapure, U.S.; Singhal, R.S. Cyclosporin A-a Review on Fermentative Production, Downstream Processing and Pharmacological Applications. Biotechnol. Adv. 2011, 29, 418–435. [Google Scholar] [CrossRef] [PubMed]
- Florea, S.; Panaccione, D.G.; Schardl, C.L. Ergot Alkaloids of the Family Clavicipitaceae. Phytopathology 2017, 107, 504–518. [Google Scholar] [CrossRef] [PubMed]
- Peñalva, M.A.; Rowlands, R.T.; Turner, G. The Optimization of Penicillin Biosynthesis in Fungi. Trends Biotechnol. 1998, 16, 483–489. [Google Scholar] [CrossRef]
- Jin, J.-M.; Lee, S.; Lee, J.; Baek, S.-R.; Kim, J.-C.; Yun, S.-H.; Park, S.-Y.; Kang, S.; Lee, Y.-W. Functional Characterization and Manipulation of the Apicidin Biosynthetic Pathway in Fusarium semitectum. Mol. Microbiol. 2010, 76, 456–466. [Google Scholar] [CrossRef]
- Ratnaweera, P.B.; de Silva, E.D.; Williams, D.E.; Andersen, R.J. Antimicrobial Activities of Endophytic Fungi Obtained from the Arid Zone Invasive Plant Opuntia dillenii and the Isolation of Equisetin, from Endophytic Fusarium sp. BMC Complement. Altern. Med. 2015, 15, 220. [Google Scholar] [CrossRef]
- Kakule, T.B.; Jadulco, R.C.; Koch, M.; Janso, J.E.; Barrows, L.R.; Schmidt, E.W. Native Promoter Strategy for High-Yielding Synthesis and Engineering of Fungal Secondary Metabolites. ACS Synth. Biol. 2015, 4, 625–633. [Google Scholar] [CrossRef]
- Niehaus, E.-M.; Díaz-Sánchez, V.; von Bargen, K.W.; Kleigrewe, K.; Humpf, H.-U.; Limón, M.C.; Tudzynski, B. Fusarins and Fusaric Acid in Fusaria. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Martín, J.-F., García-Estrada, C., Zeilinger, S., Eds.; Springer: New York, NY, USA, 2014; pp. 239–262. ISBN 978-1-4939-1190-5. [Google Scholar]
- Skellam, E. The Biosynthesis of Cytochalasans. Nat. Prod. Rep. 2017, 34, 1252–1263. [Google Scholar] [CrossRef]
- Tokuoka, M.; Seshime, Y.; Fujii, I.; Kitamoto, K.; Takahashi, T.; Koyama, Y. Identification of a Novel Polyketide Synthase-Nonribosomal Peptide Synthetase (PKS-NRPS) Gene Required for the Biosynthesis of Cyclopiazonic Acid in Aspergillus oryzae. Fungal Genet. Biol. 2008, 45, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Gil-Serna, J.; García-Díaz, M.; González-Jaén, M.T.; Vázquez, C.; Patiño, B. Description of an Orthologous Cluster of Ochratoxin A Biosynthetic Genes in Aspergillus and Penicillium Species. A Comparative Analysis. Int. J. Food Microbiol. 2018, 268, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Malir, F.; Ostry, V.; Pfohl-Leszkowicz, A.; Malir, J.; Toman, J. Ochratoxin A: 50 Years of Research. Toxins 2016, 8, 191. [Google Scholar] [CrossRef] [PubMed]
- Tudzynski, B. Gibberellin Biosynthesis in Fungi: Genes, Enzymes, Evolution, and Impact on Biotechnology. Appl. Microbiol. Biotechnol. 2005, 66, 597–611. [Google Scholar] [CrossRef] [PubMed]
- Studt, L.; Tudzynski, B. Gibberellins and the Red Pigments Bikaverin and Fusarubin. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Martín, J.-F., García-Estrada, C., Zeilinger, S., Eds.; Springer: New York, NY, USA, 2014; pp. 209–238. ISBN 978-1-4939-1190-5. [Google Scholar]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum Trichothecene Mycotoxins: Biosynthesis, Regulation, and Management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef]
- Avalos, J.; Pardo-Medina, J.; Parra-Rivero, O.; Ruger-Herreros, M.; Rodríguez-Ortiz, R.; Hornero-Méndez, D.; Limón, M.C. Carotenoid Biosynthesis in Fusarium. J. Fungi 2017, 3, 39. [Google Scholar] [CrossRef]
- Avalos, J.; Corrochano, L.M. Carotenoid Biosynthesis in Neurospora. In Neurospora: Genomics and Molecular Biology; Kasbekar, D.P., McCluskey, K., Eds.; Caister Academic Press: Poole, UK, 2013; pp. 227–241. [Google Scholar]
- Lo, H.-C.; Entwistle, R.; Guo, C.-J.; Ahuja, M.; Szewczyk, E.; Hung, J.-H.; Chiang, Y.-M.; Oakley, B.R.; Wang, C.C.C. Two Separate Gene Clusters Encode the Biosynthetic Pathway for the Meroterpenoids Austinol and Dehydroaustinol in Aspergillus nidulans. J. Am. Chem. Soc. 2012, 134, 4709–4720. [Google Scholar] [CrossRef] [PubMed]
- Mitsuguchi, H.; Seshime, Y.; Fujii, I.; Shibuya, M.; Ebizuka, Y.; Kushiro, T. Biosynthesis of Steroidal Antibiotic Fusidanes: Functional Analysis of Oxidosqualene Cyclase and Subsequent Tailoring Enzymes from Aspergillus fumigatus. J. Am. Chem. Soc. 2009, 131, 6402–6411. [Google Scholar] [CrossRef]
- Hertweck, C.; Luzhetskyy, A.; Rebets, Y.; Bechthold, A. Type II Polyketide Synthases: Gaining a Deeper Insight into Enzymatic Teamwork. Nat. Prod. Rep. 2007, 24, 162–190. [Google Scholar] [CrossRef]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone Synthase and Its Functions in Plant Resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Nosanchuk, J.D.; Stark, R.E.; Casadevall, A. Fungal Melanin: What Do We Know about Structure. Front. Microbiol. 2015, 6, 1463. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Casadevall, A. Synthesis and Assembly of Fungal Melanin. Appl. Microbiol. Biotechnol. 2012, 93, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Süssmuth, R.D.; Mainz, A. Nonribosomal Peptide Synthesis-Principles and Prospects. Angew. Chem. Int. Ed. Engl. 2017, 56, 3770–3821. [Google Scholar] [CrossRef]
- Finking, R.; Marahiel, M.A. Biosynthesis of Nonribosomal Peptides. Annu. Rev. Microbiol. 2004, 58, 453–488. [Google Scholar] [CrossRef]
- Caboche, S.; Pupin, M.; Leclère, V.; Fontaine, A.; Jacques, P.; Kucherov, G. NORINE: A Database of Nonribosomal Peptides. Nucleic Acids Res. 2008, 36, D326–D331. [Google Scholar] [CrossRef]
- Flissi, A.; Ricart, E.; Campart, C.; Chevalier, M.; Dufresne, Y.; Michalik, J.; Jacques, P.; Flahaut, C.; Lisacek, F.; Leclère, V.; et al. Norine: Update of the Nonribosomal Peptide Resource. Nucleic Acids Res. 2020, 48, D465–D469. [Google Scholar] [CrossRef] [PubMed]
- Soukup, A.A.; Keller, N.P.; Wiemann, P. Enhancing Nonribosomal Peptide Biosynthesis in Filamentous Fungi. Methods Mol. Biol. 2016, 1401, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T. Insights into the Chemical Logic and Enzymatic Machinery of NRPS Assembly Lines. Nat. Prod. Rep. 2016, 33, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Boettger, D.; Hertweck, C. Molecular Diversity Sculpted by Fungal PKS-NRPS Hybrids. Chembiochem 2013, 14, 28–42. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, J.L.; Sondergaard, T.E.; Covarelli, L.; Fuertes, P.R.; Hansen, F.T.; Frandsen, R.J.N.; Saei, W.; Lukassen, M.B.; Wimmer, R.; Nielsen, K.F.; et al. Identification of the Biosynthetic Gene Clusters for the Lipopeptides Fusaristatin A and W493 B in Fusarium graminearum and F. pseudograminearum. J. Nat. Prod. 2014, 77, 2619–2625. [Google Scholar] [CrossRef]
- Du, L.; Sánchez, C.; Shen, B. Hybrid Peptide-Polyketide Natural Products: Biosynthesis and Prospects toward Engineering Novel Molecules. Metab. Eng. 2001, 3, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Theobald, S.; Vesth, T.C.; Andersen, M.R. Genus Level Analysis of PKS-NRPS and NRPS-PKS Hybrids Reveals Their Origin in Aspergilli. BMC Genom. 2019, 20, 847. [Google Scholar] [CrossRef]
- Fisch, K.M. Biosynthesis of Natural Products by Microbial Iterative Hybrid PKS–NRPS. RSC Adv. 2013, 3, 18228. [Google Scholar] [CrossRef]
- Sacchettini, J.C.; Poulter, C.D. Creating Isoprenoid Diversity. Science 1997, 277, 1788–1789. [Google Scholar] [CrossRef]
- Lange, B.M.; Rujan, T.; Martin, W.; Croteau, R. Isoprenoid Biosynthesis: The Evolution of Two Ancient and Distinct Pathways across Genomes. Proc. Natl. Acad. Sci. USA 2000, 97, 13172–13177. [Google Scholar] [CrossRef] [PubMed]
- Heddergott, C.; Calvo, A.M.; Latgé, J.P. The Volatome of Aspergillus fumigatus. Eukaryot. Cell 2014, 13, 1014–1025. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, B.A.; Poulter, C.D. Chain Elongation in the Isoprenoid Biosynthetic Pathway. Curr. Opin. Chem. Biol. 1997, 1, 570–578. [Google Scholar] [CrossRef]
- Christianson, D.W. Unearthing the Roots of the Terpenome. Curr. Opin. Chem. Biol. 2008, 12, 141–150. [Google Scholar] [CrossRef]
- Quin, M.B.; Flynn, C.M.; Schmidt-Dannert, C. Traversing the Fungal Terpenome. Nat. Prod. Rep. 2014, 31, 1449–1473. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Dannert, C. Biosynthesis of Terpenoid Natural Products in Fungi. Adv. Biochem. Eng. Biotechnol. 2015, 148, 19–61. [Google Scholar] [CrossRef]
- Christianson, D.W. Structural Biology and Chemistry of the Terpenoid Cyclases. Chem. Rev. 2006, 106, 3412–3442. [Google Scholar] [CrossRef]
- Matsuda, Y.; Awakawa, T.; Mori, T.; Abe, I. Unusual Chemistries in Fungal Meroterpenoid Biosynthesis. Curr. Opin. Chem. Biol. 2016, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Rokas, A.; Mead, M.E.; Steenwyk, J.L.; Raja, H.A.; Oberlies, N.H. Biosynthetic Gene Clusters and the Evolution of Fungal Chemodiversity. Nat. Prod. Rep. 2020, 37, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Guo, J.; Cheng, F.; Li, S. Cytochrome P450 Enzymes in Fungal Natural Product Biosynthesis. Nat. Prod. Rep. 2021, 38, 1072–1099. [Google Scholar] [CrossRef]
- Yin, W.; Keller, N.P. Transcriptional Regulatory Elements in Fungal Secondary Metabolism. J. Microbiol. 2011, 49, 329–339. [Google Scholar] [CrossRef]
- Wiemann, P.; Willmann, A.; Straeten, M.; Kleigrewe, K.; Beyer, M.; Humpf, H.U.; Tudzynski, B. Biosynthesis of the Red Pigment Bikaverin in Fusarium fujikuroi: Genes, Their Function and Regulation. Mol. Microbiol. 2009, 72, 931–946. [Google Scholar] [CrossRef]
- Brakhage, A.A. Molecular Regulation of Beta-Lactam Biosynthesis in Filamentous Fungi. Microbiol. Mol. Biol. Rev. 1998, 62, 547–585. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal Secondary Metabolism: Regulation, Function and Drug Discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and Role of Fungal Secondary Metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A. Regulation of Fungal Secondary Metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef]
- Tudzynski, B. Nitrogen Regulation of Fungal Secondary Metabolism in Fungi. Front. Microbiol 2014, 5, 656. [Google Scholar] [CrossRef]
- Szilágyi, M.; Miskei, M.; Karányi, Z.; Lenkey, B.; Pócsi, I.; Emri, T. Transcriptome Changes Initiated by Carbon Starvation in Aspergillus nidulans. Microbiology 2013, 159, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, B.; Chávez, A.; Forero, A.; García-Huante, Y.; Romero, A.; Sánchez, M.; Rocha, D.; Sánchez, B.; Rodríguez-Sanoja, R.; Sánchez, S.; et al. Production of Microbial Secondary Metabolites: Regulation by the Carbon Source. Crit. Rev. Microbiol. 2010, 36, 146–167. [Google Scholar] [CrossRef]
- Li, B.; Chen, Y.; Tian, S. Function of PH-Dependent Transcription Factor PacC in Regulating Development, Pathogenicity, and Mycotoxin Biosynthesis of Phytopathogenic Fungi. FEBS J. 2021. [Google Scholar] [CrossRef]
- Bok, J.W.; Keller, N.P. LaeA, a Regulator of Secondary Metabolism in Aspergillus spp. Eukaryot. Cell 2004, 3, 527–535. [Google Scholar] [CrossRef]
- Bayram, O.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.-J.; Keller, N.P.; Yu, J.-H.; et al. VelB/VeA/LaeA Complex Coordinates Light Signal with Fungal Development and Secondary Metabolism. Science 2008, 320, 1504–1506. [Google Scholar] [CrossRef]
- Bayram, O.; Braus, G.H. Coordination of Secondary Metabolism and Development in Fungi: The Velvet Family of Regulatory Proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef]
- Pardo-Medina, J.; Gutiérrez, G.; Limón, M.C.; Avalos, J. Impact of the White Collar Photoreceptor WcoA on the Fusarium fujikuroi Transcriptome. Front. Microbiol. 2021, 11, 619474. [Google Scholar] [CrossRef] [PubMed]
- Tudzynski, B.; Homann, V.; Feng, B.; Marzluf, G.A. Isolation, Characterization and Disruption of the AreA Nitrogen Regulatory Gene of Gibberella fujikuroi. Mol. Gen. Genet. 1999, 261, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Woloshuk, C.P. Role of AREA, a Regulator of Nitrogen Metabolism, during Colonization of Maize Kernels and Fumonisin Biosynthesis in Fusarium verticillioides. Fungal Genet. Biol. 2008, 45, 947–953. [Google Scholar] [CrossRef]
- Wagner, D.; Wiemann, P.; Huß, K.; Brandt, U.; Fleißner, A.; Tudzynski, B. A Sensing Role of the Glutamine Synthetase in the Nitrogen Regulation Network in Fusarium fujikuroi. PLoS ONE 2013, 8, e80740. [Google Scholar] [CrossRef]
- Palmer, J.M.; Keller, N.P. Secondary Metabolism in Fungi: Does Chromosomal Location Matter? Curr. Opin. Microbiol. 2010, 13, 431–436. [Google Scholar] [CrossRef]
- Strauss, J.; Reyes-Dominguez, Y. Regulation of Secondary Metabolism by Chromatin Structure and Epigenetic Codes. Fungal Genet. Biol. 2011, 48, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Studt, L.; Rösler, S.M.; Burkhardt, I.; Arndt, B.; Freitag, M.; Humpf, H.-U.; Dickschat, J.S.; Tudzynski, B. Knock-down of the Methyltransferase Kmt6 Relieves H3K27me3 and Results in Induction of Cryptic and Otherwise Silent Secondary Metabolite Gene Clusters in Fusarium fujikuroi. Environ. Microbiol. 2016, 18, 4037–4054. [Google Scholar] [CrossRef]
- Adpressa, D.A.; Connolly, L.R.; Konkel, Z.M.; Neuhaus, G.F.; Chang, X.L.; Pierce, B.R.; Smith, K.M.; Freitag, M.; Loesgen, S. A Metabolomics-Guided Approach to Discover Fusarium graminearum Metabolites after Removal of a Repressive Histone Modification. Fungal Genet. Biol. 2019, 132, 103256. [Google Scholar] [CrossRef]
- Pfannmüller, A.; Leufken, J.; Studt, L.; Michielse, C.B.; Sieber, C.M.K.; Güldener, U.; Hawat, S.; Hippler, M.; Fufezan, C.; Tudzynski, B. Comparative Transcriptome and Proteome Analysis Reveals a Global Impact of the Nitrogen Regulators AreA and AreB on Secondary Metabolism in Fusarium fujikuroi. PLoS ONE 2017, 12, e0176194. [Google Scholar] [CrossRef] [PubMed]
- Kistler, H.C.; Broz, K. Cellular Compartmentalization of Secondary Metabolism. Front. Microbiol. 2015, 6, 68. [Google Scholar] [CrossRef] [PubMed]
- Freitas, D.F.; da Rocha, I.M.; Vieira-da-Motta, O.; de Paula Santos, C. The Role of Melanin in the Biology and Ecology of Nematophagous Fungi. J. Chem. Ecol. 2021, 47, 597–613. [Google Scholar] [CrossRef]
- Bushley, K.E.; Ripoll, D.R.; Turgeon, B.G. Module Evolution and Substrate Specificity of Fungal Nonribosomal Peptide Synthetases Involved in Siderophore Biosynthesis. BMC Evol. Biol. 2008, 8, 328. [Google Scholar] [CrossRef]
- Studt, L.; Wiemann, P.; Kleigrewe, K.; Humpf, H.-U.; Tudzynski, B. Biosynthesis of Fusarubins Accounts for Pigmentation of Fusarium fujikuroi Perithecia. Appl. Environ. Microbiol. 2012, 78, 4468–4480. [Google Scholar] [CrossRef] [PubMed]
- Frandsen, R.J.N.; Rasmussen, S.A.; Knudsen, P.B.; Uhlig, S.; Petersen, D.; Lysøe, E.; Gotfredsen, C.H.; Giese, H.; Larsen, T.O. Black Perithecial Pigmentation in Fusarium Species Is Due to the Accumulation of 5-Deoxybostrycoidin-Based Melanin. Sci. Rep. 2016, 6, 26206. [Google Scholar] [CrossRef]
- Avalos, J.; Limón, M.C. Biological Roles of Fungal Carotenoids. Curr. Genet. 2015, 61, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Medina, H.R.; Cerdá-Olmedo, E.; Al-Babili, S. Cleavage Oxygenases for the Biosynthesis of Trisporoids and Other Apocarotenoids in Phycomyces. Mol. Microbiol. 2011, 82, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Tagua, V.G.; Medina, H.R.; Martín-Domínguez, R.; Eslava, A.P.; Corrochano, L.M.; Cerdá-Olmedo, E.; Idnurm, A. A Gene for Carotene Cleavage Required for Pheromone Biosynthesis and Carotene Regulation in the Fungus Phycomyces blakesleeanus. Fungal Genet. Biol. 2012, 49, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Estrada, A.F.; Brefort, T.; Mengel, C.; Díaz-Sánchez, V.; Alder, A.; Al-Babili, S.; Avalos, J. Ustilago maydis Accumulates β-Carotene at Levels Determined by a Retinal-Forming Carotenoid Oxygenase. Fungal Genet. Biol. 2010, 46, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L.; Fang, A. The Natural Functions of Secondary Metabolites. Adv. Biochem. Eng. Biotechnol. 2000, 69, 1–39. [Google Scholar] [CrossRef]
- Fan, Y.; Liu, X.; Keyhani, N.O.; Tang, G.; Pei, Y.; Zhang, W.; Tong, S. Regulatory Cascade and Biological Activity of Beauveria bassiana Oosporein That Limits Bacterial Growth after Host Death. Proc. Natl. Acad. Sci. USA 2017, 114, E1578–E1586. [Google Scholar] [CrossRef]
- Pusztahelyi, T.; Holb, I.J.; Pócsi, I. Secondary Metabolites in Fungus-Plant Interactions. Front. Plant Sci. 2015, 6, 573. [Google Scholar] [CrossRef]
- Kai, K. Bioorganic Chemistry of Signaling Molecules in Microbial Communication. J. Pestic. Sci. 2019, 44, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Niehaus, E.-M.; von Bargen, K.W.; Espino, J.J.; Pfannmüller, A.; Humpf, H.-U.; Tudzynski, B. Characterization of the Fusaric Acid Gene Cluster in Fusarium fujikuroi. Appl. Microbiol. Biotechnol. 2014, 98, 1749–1762. [Google Scholar] [CrossRef] [PubMed]
- Motoyama, T.; Yun, C.-S.; Osada, H. Biosynthesis and Biological Function of Secondary Metabolites of the Rice Blast Fungus pyricularia Oryzae. J. Ind. Microbiol. Biotechnol. 2021, kuab058. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Schroeckh, V. Fungal Secondary Metabolites—Strategies to Activate Silent Gene Clusters. Fungal Genet. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Triggering Cryptic Natural Product Biosynthesis in Microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- López-Díaz, C.; Rahjoo, V.; Sulyok, M.; Ghionna, V.; Martín-Vicente, A.; Capilla, J.; Di Pietro, A.; López-Berges, M.S. Fusaric Acid Contributes to Virulence of Fusarium oxysporum on Plant and Mammalian Hosts. Mol. Plant. Pathol. 2018, 19, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, P.; Sieber, C.M.; von Bargen, K.W.; Studt, L.; Niehaus, E.M.; Espino, J.J.; Huss, K.; Michielse, C.B.; Albermann, S.; Wagner, D.; et al. Deciphering the Cryptic Genome: Genome-Wide Analyses of the Rice Pathogen Fusarium fujikuroi Reveal Complex Regulation of Secondary Metabolism and Novel Metabolites. PLoS Pathog. 2013, 9, e1003475. [Google Scholar] [CrossRef]
- Demain, A.L. Valuable Secondary Metabolites from Fungi. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Martín, J.-F., García-Estrada, C., Zeilinger, S., Eds.; Fungal Biology; Springer: New York, NY, USA, 2014; pp. 1–15. ISBN 978-1-4939-1190-5. [Google Scholar]
- Wu, F.; Groopman, J.D.; Pestka, J.J. Public Health Impacts of Foodborne Mycotoxins. Annu. Rev. Food Sci. Technol. 2014, 5, 351–372. [Google Scholar] [CrossRef]
- Marin, S.; Ramos, A.J.; Cano-Sancho, G.; Sanchis, V. Mycotoxins: Occurrence, Toxicology, and Exposure Assessment. Food Chem. Toxicol. 2013, 60, 218–237. [Google Scholar] [CrossRef]
- Schmitt, E.K.; Hoff, B.; Kück, U. Regulation of Cephalosporin Biosynthesis. Adv. Biochem. Eng. Biotechnol. 2004, 88, 1–43. [Google Scholar] [CrossRef]
- Petersen, A.B.; Rønnest, M.H.; Larsen, T.O.; Clausen, M.H. The Chemistry of Griseofulvin. Chem. Rev. 2014, 114, 12088–12107. [Google Scholar] [CrossRef]
- Avalos, J.; Nordzieke, S.; Parra, O.; Pardo-Medina, J.; Limón, M.C. Carotenoid Production by Filamentous Fungi and Yeasts. In Biotechnology of Yeasts and Filamentous Fungi; Sibirny, A.A., Ed.; Springer: Cham, Switzerland, 2017; pp. 225–279. ISBN 978-3-319-58828-5. [Google Scholar]
- Amna, T.; Amina, M.; Sharma, P.R.; Puri, S.C.; Al-Youssef, H.M.; Al-Taweel, A.M.; Qazi, G.N. Effect of Precursors Feeding and Media Manipulation on Production of Novel Anticancer Pro-Drug Camptothecin from Endophytic Fungus. Braz. J. Microbiol. 2012, 43, 1476–1490. [Google Scholar] [CrossRef]
- Boruta, T. Uncovering the Repertoire of Fungal Secondary Metabolites: From Fleming’s Laboratory to the International Space Station. Bioengineered 2018, 9, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Sáiz, M.; de la Fuente, J.L.; Barredo, J.L. Xanthophyllomyces dendrorhous for the Industrial Production of Astaxanthin. Appl. Microbiol. Biotechnol. 2010, 88, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Shao, Y.; Chen, F. Monascus Pigments. Appl. Microbiol. Biotechnol. 2012, 96, 1421–1440. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.I.; Miller, J.D. A Concise History of Mycotoxin Research. J. Agric. Food Chem. 2017, 65, 7021–7033. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Ghazali, F.M.; Mahyudin, N.A.; Samsudin, N.I.P. Aflatoxin Biosynthesis, Genetic Regulation, Toxicity, and Control Strategies: A Review. J. Fungi 2021, 7, 606. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Proctor, R.H. Molecular Biology of Fusarium Mycotoxins. Int. J. Food. Microbiol. 2007, 119, 47–50. [Google Scholar] [CrossRef]
- Zhu, B.; Jeffrey, A.M. Fusarin C: Isolation and Identification of Two Microsomal Metabolites. Chem. Res. Toxicol. 1993, 6, 97–101. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Andersen, B.; Thrane, U. The Use of Secondary Metabolite Profiling in Chemotaxonomy of Filamentous Fungi. Mycol. Res. 2008, 112, 231–240. [Google Scholar] [CrossRef]
- Molnár, K.; Farkas, E. Current Results on Biological Activities of Lichen Secondary Metabolites: A Review. Z. Naturforsch C. J. Biosci. 2010, 65, 157–173. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.-H.; Chollet-Krugler, M.; Gouault, N.; Tomasi, S. UV-Protectant Metabolites from Lichens and Their Symbiotic Partners. Nat. Prod. Rep. 2013, 30, 1490–1508. [Google Scholar] [CrossRef]
- Olivier-Jimenez, D.; Chollet-Krugler, M.; Rondeau, D.; Beniddir, M.A.; Ferron, S.; Delhaye, T.; Allard, P.-M.; Wolfender, J.-L.; Sipman, H.J.M.; Lücking, R.; et al. A Database of High-Resolution MS/MS Spectra for Lichen Metabolites. Sci. Data 2019, 6, 294. [Google Scholar] [CrossRef]
- Schueffler, A.; Anke, T. Fungal Natural Products in Research and Development. Nat. Prod. Rep. 2014, 31, 1425–1448. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yu, R.; Bai, X.; Wang, H.; Zhang, H. Fusarium: A Treasure Trove of Bioactive Secondary Metabolites. Nat. Prod. Rep. 2020, 37, 1568–1588. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; de Rond, T.; Moore, B.S. Mining Genomes to Illuminate the Specialized Chemistry of Life. Nat. Rev. Genet. 2021, 22, 553–571. [Google Scholar] [CrossRef]
- Khaldi, N.; Seifuddin, F.T.; Turner, G.; Haft, D.; Nierman, W.C.; Wolfe, K.H.; Fedorova, N.D. SMURF: Genomic Mapping of Fungal Secondary Metabolite Clusters. Fungal Genet. Biol. 2010, 47, 736–741. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Umemura, M.; Koike, H.; Machida, M. Motif-Independent de Novo Detection of Secondary Metabolite Gene Clusters-toward Identification from Filamentous Fungi. Front. Microbiol. 2015, 6, 371. [Google Scholar] [CrossRef]
- Winter, J.M.; Behnken, S.; Hertweck, C. Genomics-Inspired Discovery of Natural Products. Curr. Opin. Chem. Biol. 2011, 15, 22–31. [Google Scholar] [CrossRef]
- Wiemann, P.; Keller, N.P. Strategies for Mining Fungal Natural Products. J. Ind. Microbiol. Biotechnol. 2014, 41, 301–313. [Google Scholar] [CrossRef]
- Kjærbølling, I.; Mortensen, U.H.; Vesth, T.; Andersen, M.R. Strategies to Establish the Link between Biosynthetic Gene Clusters and Secondary Metabolites. Fungal. Genet. Biol. 2019, 130, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.F.; Somoza, A.D.; Keller, N.P.; Wang, C.C.C. Advances in Aspergillus Secondary Metabolite Research in the Post-Genomic Era. Nat. Prod. Rep. 2012, 29, 351–371. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.C.; Grijseels, S.; Prigent, S.; Ji, B.; Dainat, J.; Nielsen, K.F.; Frisvad, J.C.; Workman, M.; Nielsen, J. Global Analysis of Biosynthetic Gene Clusters Reveals Vast Potential of Secondary Metabolite Production in Penicillium Species. Nat. Microbiol. 2017, 2, 17044. [Google Scholar] [CrossRef] [PubMed]
- Hoogendoorn, K.; Barra, L.; Waalwijk, C.; Dickschat, J.S.; van der Lee, T.A.J.; Medema, M.H. Evolution and Diversity of Biosynthetic Gene Clusters in Fusarium. Front. Microbiol. 2018, 9, 1158. [Google Scholar] [CrossRef]
- Mattern, D.J.; Valiante, V.; Unkles, S.E.; Brakhage, A.A. Synthetic Biology of Fungal Natural Products. Front. Microbiol. 2015, 6, 775. [Google Scholar] [CrossRef] [PubMed]
- Kornfuehrer, T.; Eustáquio, A.S. Diversification of Polyketide Structures via Synthase Engineering. Medchemcomm 2019, 10, 1256–1272. [Google Scholar] [CrossRef]
- Hwang, S.; Lee, N.; Cho, S.; Palsson, B.; Cho, B.-K. Repurposing Modular Polyketide Synthases and Non-Ribosomal Peptide Synthetases for Novel Chemical Biosynthesis. Front. Mol. Biosci. 2020, 7, 87. [Google Scholar] [CrossRef]

| Chemical Family | Metabolite | Function/Activity | Representative Producing Genera | Reference |
|---|---|---|---|---|
| Polyketides (PKs) | Fumonisin B1 | Mycotoxin | Fusarium | [11] |
| Lovastatin | HMG-CoA reductase inhibitor | Aspergillus | [12] | |
| Aflatoxin | Mycotoxin | Aspergillus | [13] | |
| Bikaverin | Antibiotic (protozoa) | Fusarium | [14] | |
| Zearalenone | Mycotoxin (estrogenic) | Fusarium | [10] | |
| Non ribosomal peptides (NRPs) | Enniatin B | Mycotoxin (cytotoxic) | Fusarium | [15] |
| Cyclosporine A | Immunosuppressant | Tolypocladium | [16] | |
| Ergotamine | Ergot alkaloid | Claviceps | [17] | |
| Penicillin G | Antibiotic (bacteria) | Penicillium | [18] | |
| Apicidin | Histone deacetylase inhibitor | Fusarium | [19] | |
| Hybrid NRP/PKs | Equisetin | Antibiotic (bacteria) | Fusarium | [20,21] |
| Fusarin C | Mycotoxin | Fusarium | [22] | |
| Cytochalasin | Actin inhibitor | Penicillium, Chaetomium | [23] | |
| Cyclopiazonic acid | Mycotoxin | Aspergillus, Penicillium | [24] | |
| Ochratoxin A | Mycotoxin | Aspergillus, Penicillium | [25,26] | |
| Terpenoids | Gibberellic acid (GA3) | Plant hormone | Fusarium | [27,28] |
| Deoxynivalenol | Mycotoxin | Fusarium | [29] | |
| Neurosporaxanthin | Carotenoid pigment | Neurospora, Fusarium | [30,31] | |
| Austinol | Unknown | Aspergillus | [32] | |
| Helvolic acid | Antibiotic (bacteria) | Aspergillus | [33] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avalos, J.; Limón, M.C. Fungal Secondary Metabolism. Encyclopedia 2022, 2, 1-13. https://doi.org/10.3390/encyclopedia2010001
Avalos J, Limón MC. Fungal Secondary Metabolism. Encyclopedia. 2022; 2(1):1-13. https://doi.org/10.3390/encyclopedia2010001
Chicago/Turabian StyleAvalos, Javier, and M. Carmen Limón. 2022. "Fungal Secondary Metabolism" Encyclopedia 2, no. 1: 1-13. https://doi.org/10.3390/encyclopedia2010001