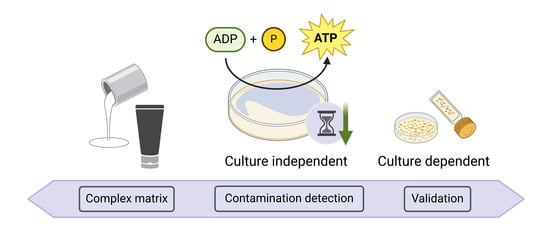
Breaking the Mold: Towards Rapid and Cost-Effective Microbial Contamination Detection in Paints and Cosmetics Using ATP-Bioluminescence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Specimens
2.2. Test Organisms, Growth Conditions, and Preparation of Pure and Mixed Inocula
2.3. Calibration Curves
2.4. Matrix Spiking
2.5. Test Protocol and Method Validation
2.5.1. Standard Plate Count
2.5.2. ATP Bioluminescence Assay
2.5.3. Dipslides
2.6. Statistical Analyses
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jasson, V.; Jacxsens, L.; Luning, P.; Rajkovic, A.; Uyttendaele, M. Alternative microbial methods: An overview and selection criteria. Food Microbiol. 2010, 27, 710–730. [Google Scholar] [CrossRef]
- Bottari, B.; Santarelli, M.; Neviani, E. Determination of microbial load for different beverages and foodstuff by assessment of intracellular ATP. Trends Food Sci. 2015, 44, 36–48. [Google Scholar] [CrossRef]
- Monica, S.; Bancalari, E.; Castellone, V.; Rijkx, J.; Wirth, S.; Jahns, A.; Bottari, B. ATP bioluminescence for rapid and selective detection of bacteria and yeasts in wine. Appl. Sci. 2021, 11, 4953. [Google Scholar] [CrossRef]
- Emerson, J.B.; Adams, R.I.; Román, C.M.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 2017, 5, 1–23. [Google Scholar] [CrossRef]
- García-Nicolás, M.; Arroyo-Manzanares, N.; Arce, L.; Hernández-Córdoba, M.; Viñas, P. Headspace gas chromatography coupled to mass spectrometry and ion mobility spectrometry: Classification of virgin olive oils as a study case. Foods 2020, 9, 1288. [Google Scholar] [CrossRef]
- Alvarez-Rivera, G.; De Miguel, T.; Llompart, M.; Garcia-Jares, C.; Villa, T.G.; Lores, M. A novel outlook on detecting microbial contamination in cosmetic products: Analysis of biomarker volatile compounds by solid-phase microextraction gas chromatography-mass spectrometry. Anal. Methods 2013, 5, 384–393. [Google Scholar] [CrossRef]
- Celeiro, M.; Varela, E.; Rodriguez, R.; Penedo, M.; Lores, M. Tracking bacterial spoilage in cosmetics by a new bioanalytical approach: API-SPME-GC-MS to monitor MVOCs. Cosmetics 2020, 7, 38. [Google Scholar] [CrossRef]
- Ahn, Y.; Gibson, B.; Williams, A.; Alusta, P.; Buzatu, D.A.; Lee, Y.J.; LiPuma, J.J.; Hussong, D.; Marasa, B.; Cerniglia, C.E. A comparison of culture-based, real-time PCR, droplet digital PCR and flow cytometric methods for the detection of Burkholderia cepacia complex in nuclease-free water and antiseptics. J. Ind. Microbiol. Biotech. 2020, 47, 475–484. [Google Scholar] [CrossRef]
- Cao, Y.; Yu, M.; Dong, G.; Chen, B.; Zhang, B. Digital PCR as an emerging tool for monitoring of microbial biodegradation. Molecules 2020, 25, 706. [Google Scholar] [CrossRef]
- Mutschlechner, M.; Walter, A.; Bach, K.; Schöbel, H. Beyond Cultivation: Combining Culture-Dependent and Culture-Independent Techniques to Identify Bacteria Involved in Paint Spoilage. Coatings 2023, 13, 1055. [Google Scholar] [CrossRef]
- Sanna, T.; Dallolio, L.; Raggi, A.; Mazzetti, M.; Lorusso, G.; Zanni, A.; Farruggia, P.; Leoni, E. ATP bioluminescence assay for evaluating cleaning practices in operating theatres: Applicability and limitations. BMC Infect. Dis. 2018, 18, 1–7. [Google Scholar] [CrossRef]
- Lane, K.; McLandsborough, L.A.; Autio, W.R.; Kinchla, A.J. Efficacy of ATP monitoring for measuring organic matter on postharvest food contact surfaces. J. Food Prot. 2020, 83, 1829–1837. [Google Scholar] [CrossRef]
- Martens, R. Estimation of ATP in soil: Extraction methods and calculation of extraction efficiency. Soil Biol. Biochem. 2001, 33, 973–982. [Google Scholar] [CrossRef]
- Braissant, O.; Astasov-Frauenhoffer, M.; Waltimo, T.; Bonkat, G. A review of methods to determine viability, vitality, and metabolic rates in microbiology. Front. Microbiol. 2020, 11, 547458. [Google Scholar] [CrossRef]
- Pandey, P.; Kiran, U.V. Degradation of paints and its microbial effect on health and environment. J. Crit. Rev. 2020, 7, 4879–4884. [Google Scholar]
- Jiménez-López, A.M.; Hincapié-Llanos, G.A. Identification of factors affecting the reduction of VOC emissions in the paint industry: Systematic literature review-SLR. Prog. Org. Coat. 2022, 170, 106945. [Google Scholar] [CrossRef]
- La Rosa, F.R.; Giese, E.C.; Dekker RF, H.; Pelayo, J.S.; Barbosa, A.M. Microbiological contamination of water-based paints from an industry in the state of Paraná, Brazil. Braz. J. Environ. Sci. (RBCIAMB) 2008, 29, 85–92. [Google Scholar]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef]
- Luo, J.; Liu, X.; Tian, Q.; Yue, W.; Zeng, J.; Chen, G.; Cai, X. Disposable bioluminescence-based biosensor for detection of bacterial count in food. Anal. Biochem. 2009, 394, 1–6. [Google Scholar] [CrossRef]
- Venkateswaran, K.; Hattori, N.; La Duc, M.T.; Kern, R. ATP as a biomarker of viable microorganisms in clean-room facilities. J. Microbiol. Methods 2003, 52, 367–377. [Google Scholar] [CrossRef]
- Montanez, G.; Passman, F.; Machtiger, N.; Montemayor, R.; Whalen, P. Bioluminescence testing for rapid detection of microbial contamination in aqueous polymer emulsions. Int. Biodeterior. Biodegrad. 2016, 114, 216–221. [Google Scholar] [CrossRef]
- Connolly, P.; Bloomfield, S.F.; Denyer, S.P. A study of the use of rapid methods for preservative efficacy testing of pharmaceuticals and cosmetics. J. Appl. Bacteriol. 1993, 75, 456–462. [Google Scholar] [CrossRef]
- Narsaiah, K.; Jha, S.N.; Jaiswal, P.; Singh, A.K.; Gupta, M.; Bhardwaj, R. Estimation of total bacteria on mango surface by using ATP bioluminescence. Sci. Hortic. 2012, 146, 159–163. [Google Scholar] [CrossRef]
- Bakke, M. A comprehensive analysis of ATP tests: Practical use and recent progress in the total adenylate test for the effective monitoring of hygiene. J. Food Prot. 2022, 85, 1079–1095. [Google Scholar] [CrossRef]
- Phillips, Z.E.V.; Strauch, M.A. Bacillus subtilis sporulation and stationary phase gene expression. Cell. Mol. Life Sci. 2002, 59, 392–402. [Google Scholar] [CrossRef]
- Hironaka, I.; Iwase, T.; Sugimoto, S.; Okuda, K.I.; Tajima, A.; Yanaga, K.; Mizunoe, Y. Glucose triggers ATP secretion from bacteria in a growth-phase-dependent manner. Appl. Environ. Microbiol. 2013, 79, 2328–2335. [Google Scholar] [CrossRef]
- Fajardo-Cavazos, P.; Nicholson, W. Bacillus endospores isolated from granite: Close molecular relationships to globally distributed Bacillus spp. from endolithic and extreme environments. Appl. Environ. Microbiol. 2006, 72, 2856–2863. [Google Scholar] [CrossRef]
- Nicholson, W.L.; Fajardo-Cavazos, P.; Rebeil, R.; Slieman, T.A.; Riesenman, P.J.; Law, J.F.; Xue, Y. Bacterial endospores and their significance in stress resistance. Antonie Van Leeuwenhoek 2002, 81, 27–32. [Google Scholar] [CrossRef]
- Mempin, R.; Tran, H.; Chen, C.; Gong, H.; Kim Ho, K.; Lu, S. Release of extracellular ATP by bacteria during growth. BMC Microbiol. 2013, 13, 1–13. [Google Scholar] [CrossRef]
- Nescerecka, A.; Juhna, T.; Hammes, F. Behavior and stability of adenosine triphosphate (ATP) during chlorine disinfection. Water Res. 2016, 101, 490–497. [Google Scholar] [CrossRef]
- DeLuca, M.; Wannlund, J.; McElroy, W.D. Factors affecting the kinetics of light emission from crude and purified firefly luciferase. Anal. Biochem. 1979, 95, 194–198. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, X.; Sun, W.; Nie, T.; Quanquin, N.; Sun, Y. Escherichia coli increases its ATP concentration in weakly acidic environments principally through the glycolytic pathway. Genes 2020, 11, 991. [Google Scholar] [CrossRef]
- Paciello, L.; Falco, F.C.; Landi, C.; Parascandola, P. Strengths and weaknesses in the determination of Saccharomyces cerevisiae cell viability by ATP-based bioluminescence assay. Enzyme Microb. Technol. 2013, 52, 157–162. [Google Scholar] [CrossRef]
- Phulpoto, A.H.; Maitlo, M.A.; Kanhar, N.A. Culture-dependent to culture-independent approaches for the bioremediation of paints: A review. Int. J. Environ. Sci. Technol. 2021, 18, 241–262. [Google Scholar] [CrossRef]
- Halla, N.; Fernandes, I.P.; Heleno, S.A.; Costa, P.; Boucherit-Otmani, Z.; Boucherit, K.; Rodrigues, A.E.; Ferreira, I.C.F.R.; Barreiro, M.F. Cosmetics preservation: A review on present strategies. Molecules 2018, 23, 1571. [Google Scholar] [CrossRef]
- Cappitelli, F.; Principi, P.; Pedrazzani, R.; Toniolo, L.; Sorlini, C. Bacterial and fungal deterioration of the Milan Cathedral marble treated with protective synthetic resins. Sci. Total Environ. 2007, 385, 172–181. [Google Scholar] [CrossRef]
- Olayide, O.F.; Ayomikun, K.E.; Temitope, F.O. A first report on the identification of a novel archaea, Methanospirillum lacunae from spoilt paints in Lagos, Nigeria using a metagenomic approach. Sci. Afr. 2022, 15, e01029. [Google Scholar] [CrossRef]
- Obidi, O.F.; Aboaba, O.O.; Makanjuola, M.S.; Nwachukwu SC, U. Microbial evaluation and deterioration of paints and paint-products. J. Environ. Biol. 2009, 30, 835. [Google Scholar]
- Bernauer, U.; Bodin, L.; Chaudhry, Q.; Coenraads, P.J.; Dusinska, M.; Ezendam, J.; Gaffet, E.; Galli, C.L.; Granum, B.; Panteri, E.; et al. The SCCS Notes of Guidance for the testing of cosmetic ingredients and their safety evaluation, 11th revision, 30–31 March 2021, SCCS/1628/21. Regul. Toxicol. Pharmacol. 2021, 127, 105052. [Google Scholar] [CrossRef]
- Turner, D.E.; Daugherity, E.K.; Altier, C.; Maurer, K.J. Efficacy and limitations of an ATP-based monitoring system. Lab. Anim. Sci. 2010, 49, 190–195. [Google Scholar]
- Kawabe, S.; Uchiho, Y. Development of a highly sensitive microplate luminometer using ATP bioluminescence. Luminescence 2020, 35, 1195–1198. [Google Scholar] [CrossRef] [PubMed]
- Tehri, N.; Kumar, N.; Raghu, H.V.; Vashishth, A. Biomarkers of bacterial spore germination. Ann. Microbiol. 2018, 68, 513–523. [Google Scholar] [CrossRef]
- Ghosh, S.; Korza, G.; Maciejewski, M.; Setlow, P. Analysis of metabolism in dormant spores of Bacillus species by 31P nuclear magnetic resonance analysis of low-molecular-weight compounds. J. Bacteriol. 2015, 197, 992–1001. [Google Scholar] [CrossRef] [PubMed]




| Target Concentration | Actual Spiking Concentration (CFU mL−1) |
|---|---|
| 107 | 4.35 × 106–1.99 × 107 |
| 106 | 6.97 × 105–1.59 × 106 |
| 105 | 5.02 × 104–2.99 × 105 |
| 104 | 6.53 × 104–7.67 × 104 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mutschlechner, M.; Chisté, D.; Schöbel, H. Breaking the Mold: Towards Rapid and Cost-Effective Microbial Contamination Detection in Paints and Cosmetics Using ATP-Bioluminescence. Appl. Microbiol. 2024, 4, 582-593. https://doi.org/10.3390/applmicrobiol4020040
Mutschlechner M, Chisté D, Schöbel H. Breaking the Mold: Towards Rapid and Cost-Effective Microbial Contamination Detection in Paints and Cosmetics Using ATP-Bioluminescence. Applied Microbiology. 2024; 4(2):582-593. https://doi.org/10.3390/applmicrobiol4020040
Chicago/Turabian StyleMutschlechner, Mira, Daniela Chisté, and Harald Schöbel. 2024. "Breaking the Mold: Towards Rapid and Cost-Effective Microbial Contamination Detection in Paints and Cosmetics Using ATP-Bioluminescence" Applied Microbiology 4, no. 2: 582-593. https://doi.org/10.3390/applmicrobiol4020040