1. Introduction
Medical imaging modalities such as X-ray, CT, and MRI are often used as a non-invasive procedure to obtain physiological data [
1]. Frequently, clinicians use these data for the early detection and diagnosis of diseases such as cancer and certain cardiovascular conditions [
2,
3]. However, variations in the pathology of diseases and the physiology of patients can make it difficult for human clinicians to reliably deliver accurate diagnoses [
4]. Additionally, although advancements in technology have made medical imaging data more widely available, their development has resulted in more complex datasets [
5,
6]. To address these issues, extensive research has gone into the creation of computer-aided diagnosis (CAD) tools to improve the quality of diagnoses by supporting the clinician’s decision-making process [
7,
8].
CAD tools often implement artificial intelligence (AI) or machine learning (ML) models for their ability to learn patterns from data [
9]. An AI/ML classifier is a model that can be trained using previously collected clinical data to predict whether data from a new sample indicate a positive or negative diagnosis. While AI/ML diagnostic classifiers have achieved good performance for diagnosing certain diseases in the literature, they have not seen widespread clinical implementation [
10,
11,
12]. This is partially because it has been shown that AI/ML classifiers can perform poorly when the information contained within the data is complex, i.e., if the data are heterogeneous or noisy [
13]. This is especially problematic as differences in techniques, clinical practices, and testing equipment can all be sources of noise in a dataset.
There have been many case studies demonstrating improved reliability for specific classifiers for certain applications. Mardani et al. developed a generative adversarial network (GAN) with an affine projection operator to remove aliasing artifacts from MRI images. This model was shown to be robust to even extreme forms of Gaussian noise [
14]. Janizek et al. developed a model that improved generalization between clinics by alternating training between an adversary attempting to predict adversarial samples and a classifier trying to fool the adversary [
15]. Another application to cancer MRI images used deep feature extraction to find a feature set that achieved high performance across 13 convolutional neural networks and was generalizable [
16]. Gulzar and Khan were able to use a TransUNet model for a skin cancer segmentation model robust to distortions [
17]. Further research has used adversarial defense methods such as MedRDF, non-local codex encoder (NLCE) modules, and kernel density adversary detection to improve the robustness of deep medical imaging classifiers to adversarial noise [
18,
19,
20]. There are no widespread guidelines used to evaluate model robustness [
21,
22,
23]. Therefore, the robustness of classifiers, i.e., the ability that allows translating the predictions to new, potentially noisy, data, remains a barrier to entry for the clinical implementation of AI/ML diagnostic models.
Previous studies have shown that even classifiers with good performance can lack robustness [
24,
25]. This lack of stability amongst classifiers has led to increased efforts to characterize the robustness of imaging-based classifiers. To this end, many researchers have focused on creating “benchmark sets” or test sets that contain perturbed/augmented images [
26,
27,
28]. Robustness is then typically measured by how well a given classifier, usually trained with relatively low-noise data, predicts the benchmark set images. Recently, studies have shown that adversarial training or data augmentation can potentially improve the robustness of classifiers applied to medical imaging studies [
29,
30]. One example of this is ROOD-MRI, which enables researchers to evaluate the sensitivity of their MRI segmentation model to out-of-distribution and corrupted samples [
31]. Another example is CAMELYON17-WILDS, which serves as a benchmark set for in-the-wild distribution shifts, e.g., heterogeneity between hospitals, for brain tumor segmentation [
32]. Furthermore, benchmarks have been developed to evaluate the adversarial robustness of classifiers pertaining to skin cancer classification and chest X-ray images [
26,
29].
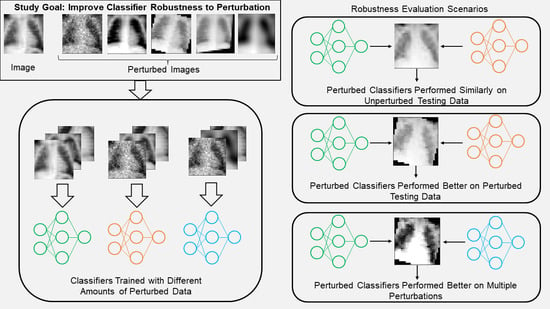
While data augmentation has been effectively used to increase classifier robustness to perturbation, there are still gaps in the literature that prevent these from being used for diagnostic tests [
33]. First, to the best of the authors’ knowledge, there is no research that explores the effect on a classifier’s robustness when different amounts of the training/test images are perturbed simultaneously. As such, dataset heterogeneity such as measurements at multiple clinical sites or using different equipment is often ignored. Second, most studies add perturbed images to the training set with the goal of improving classifier robustness, rather than replacing the original images to determine how the classifier changes what it learns [
34,
35]. Finally, there is only very limited research regarding multiple perturbations, and none that addresses the scenario where not all images receive the same combination of perturbations [
36]. To fill this gap, the contributions of this study are to (1) show that deep learning classifiers trained with medical images augmented by common perturbations can improve performance on perturbed images without necessarily sacrificing performance on unperturbed data, (2) demonstrate a perturbation scheme that allows researcher to evaluate the extent to which a classifier is robust to perturbed data, and (3) determine if classifiers trained with sets of different perturbations will perform well on images simultaneously perturbed by multiple perturbations.
The next section (
Section 2) describes the details of the procedure used in this study, as well as the datasets and types of noise used.
Section 3 presents the results of applying this procedure to two example datasets.
Section 4 discusses the results and interprets the findings. Finally,
Section 5 reflects on the overall implications of this study.
3. Results
The first step of this work was to obtain the baseline performance of each classifier by using them to predict the unperturbed test set data.
Figure 5 shows the error rate (percent of images incorrectly classified) of each classifier
fpn for all combinations of
p and
n (when
p = 0; this is the same as the unperturbed classifier). The exact numeric results for this graph can be found in
Table A1. The plotted error rates were averaged over 10 different neural network architectures. The unperturbed classifier achieved approximately a 15.3% error rate on the test data, which demonstrates that the classifier can learn the underlying data distribution. There was no noticeable decrease in the accuracy for the classifiers perturbed with 20% of the data (
f20n). There was also a minimal increase in the error rate (<0.03) for all classifiers aside from those trained with 80–100% of the training images perturbed. Intuitively, this makes sense, because the classifier was mostly trained with perturbed data and was, thus, not learning as much of the unperturbed data distribution.
Table A2 and
Table A3 contain the precision and recall of each classifier when predicting unperturbed data. Overall, the recall for all classifiers remained quite high (>0.9), while the precision was more varied (0.676–0.886), indicating that the classifiers had higher false positive rates than false negative rates on unperturbed data.
The behavior of classifiers on perturbed test set images was also analyzed for five classifier types: unperturbed,
f20n (classifier perturbed with 20% perturbed training images),
f60n (classifier perturbed with 60% perturbed training images),
f100n (classifier perturbed with 100% perturbed training images), and the SPSA adversarial classifier. As per
Figure 4, for each of the 10 network architectures, classifiers were re-trained using only one of the perturbations at a time. The average error rates of the 10 deep classifier architectures for each classifier type on each perturbation are presented in
Figure 6. The corresponding numeric data are presented in
Table A4. The unperturbed classifier’s error rate increased as the percent of perturbed images in the test set increased for rotation, tilt, and blur and, to a lesser extent, Gaussian noise. One notable result was that the unperturbed classifier’s performance did not change much at any percentage of contrast-perturbed images. The adversarial classifier exhibited similar performance trends on the rotation-, tilt-, and blur-perturbed test data, indicating that the adversarial classifier was not capable of learning these perturbations. The adversarial classifier performed better than the unperturbed classifier on noise-perturbed and contrast-perturbed data, which may indicate similarities between SPSA adversarial noise and how these perturbations affect the data.
In addition to accuracy, it was also important to measure other metrics to obtain a more comprehensive view of the classifiers’ behavior. Precision and recall were also measured, and these data are presented in
Table A6 and
Table A7. Overall, with more perturbed test data, the precision of the unperturbed classifier tended to decrease and the recall stayed high, which may indicate that the noise caused the classifier to favor negative samples over positive samples when perturbations were introduced. The changes in precision are reflective of the changes in the error rate, as the unperturbed classifier showed decreases in precision similar to its increases in the error rate for rotation-, tilt-, and blur-perturbed images. This is in contrast to the perturbed classifiers, where precision are affected less severely.
Each of the perturbed classifiers showed very little difference in performance from any of the test set scenarios, with the exception of the performance of
f100n on contrast-perturbed data. Otherwise, all of the perturbed classifiers mostly outperformed the unperturbed and adversarial classifiers. All perturbed classifiers except
f100contrast,
f60contrast, and
f60noise had a significantly lower error rate (better performance) than the unperturbed classifier for most amounts of perturbed test data. The specific
p-values computed via the Wilcoxon Signed Rank test comparing the average performance of each of the 10 unperturbed classifiers to each of the 10 perturbed classifiers are presented in
Table A7. These data show that, in addition to being able to perform well on the unperturbed test data, the performance also significantly improved on perturbed test data for most perturbed classifiers. Furthermore, there was little difference between the performance of each of the perturbed classifiers, demonstrating that there was not a linear relationship between the percentage of perturbed training images and the decrease in the performance of partially perturbed test sets. The computation of the average structural similarity index metric (SSIM) between unperturbed test sets and perturbed test sets also revealed that there was no linear relationship between perturbation intensity and classifier performance. These data are presented in
Table A8. Essentially, while the blur perturbation had a more significant impact on classifier performance than noise or contrast, the average SSIM of blur-perturbed test set images was 0.827, whereas the noise and contrast were actually more intense perturbations with SSIMs of 0.678 and 0.632.
In a separate investigation, evaluating how training on perturbed datasets that simultaneously involved how multiple perturbations will affect the robustness of the developed classifier was also looked at. This task involved both images from the PneumoniaMNIST and, separately, from the BUSI dataset. This investigation was performed by training 10 neural networks (of different architectures) on the unperturbed training data for both the X-ray and ultrasound datasets (20 total). Then, 10% of the training dataset was perturbed by each of the five perturbations used in this study, leaving 50% unperturbed, and each classifier was re-trained (by shuffling the previous weights) with these data to obtain another 20 total classifiers trained using somewhat perturbed training data. For each test sample, each perturbation had a 50% chance of being chosen to be applied to that sample, meaning each sample could be perturbed by some, all, or none of the perturbations.
Figure 7A shows the performance of the perturbed and unperturbed versions of classifiers from each network architecture trained on PneumoniaMNIST on this version of the perturbed dataset. The perturbed classifiers performed significantly better based on the Wilcoxon Signed Rank
p-value (0.000977) between the perturbed classifier error rates and unperturbed classifier error rates. On average, the perturbed classifiers had an error rate 0.0986 lower, or an improvement of 9.86%, compared to their unperturbed counterparts for the PneumoniaMNIST dataset.
Figure 7B shows this same behavior for the classifiers trained with BUSI, with an error rate decrease of 0.0515 between perturbed and unperturbed classifiers. Again, the perturbed classifiers performed significantly better (
p = 0.00684) than their unperturbed counterparts on the BUSI data. The exact error rates of each classifier’s predictions on images subject to multiple perturbations is presented in
Table A9. The error rates for each network architecture are included in
Table A7.
4. Discussion
The main goal of this study was to investigate a procedure for obtaining robust medical imaging classifiers by using perturbations on both the training and test set images. Essentially, the stability of a classifier’s performance on both perturbed and unperturbed data was evaluated. The given classifier for this study was simulated by training a series of 10 convolutional neural networks on an example dataset. Each of these networks was then passed through the proposed procedure, and their error rates were averaged over all 10 networks.
Given a classifier
f and a dataset, we evaluated the robustness of this classifier by comparing it to classifiers that were trained on perturbed training data where some percentage of images were perturbed i.e., where one would expect the classifier to perform well. In
Figure 6C–E, the unperturbed classifier
f performs worse (i.e., increases the error rate) as the percentage of test images that are perturbed increases. By perturbing even a small amount of the training set and re-training the classifier weights, significant improvements in classifier performance can be achieved on data perturbed by tilt, blur, and rotation. This implies that the original classifier was not learning information that would help it classify perturbed images, as even a small amount of perturbation in the training set resulted in noticeable improvements in performance. As such, this difference in performance between the perturbed classifier and the original classifier can be viewed as an indicator of the given classifier’s robustness (or lack thereof). Therefore, based on this procedure, the classifier
f would not be considered robust to these perturbations. Conversely, although the classifier trained with Gaussian-noise-perturbed and contrast-perturbed data performed better than the unperturbed classifier, the original classifier still showed relatively high performance and did not decrease meaningfully in performance with respect to the percentage of test set images perturbed. While the perturbed classifier may technically be more robust than the given classifier to these perturbations, this demonstrates that the original classifier was still somewhat stable given changes in contrast and noise as it still performed well (maintains < 20% error rate). Ultimately, this shows that it is important to perturb some amount of both the training and test set images to generate an increase in robustness as a trade-off for a minimal loss in accuracy.
Another finding of this work is that a robust classifier can perform well on both perturbed and unperturbed test data. For example, while the error rate of the given classifier
f was high in response to 60–100% rotation-, tilt-, and blur-perturbed test data, the error rate was still acceptable (<20%) with 20% of the test images perturbed. Therefore, it may be possible to determine just how much noise a classifier can handle before it is no longer useful. For instance, if 20% of the data are measured in a different clinic or using a different machine than the rest of the data, then the classifier may still perform reasonably. However, if half the data include this different distribution, it may not. Furthermore, the difference between the perturbed classifier’s performance and the unperturbed classifier’s performance was similar for each perturbation, as can be seen in
Figure 6. However, as previously stated, as more of the data are perturbed, it is clear that the original classifier was much less robust to rotation, tilt, and blur perturbations than Gaussian noise and contrast. Therefore, a classifier may be robust to perturbations when only a small amount of the data are perturbed; however, it cannot be said whether this robustness will be maintained or diminish based on this one performance test.
If the sample size allows, perturbing a portion of data may improve the robustness of the classifier. For example, f20rotation performed similarly on the unperturbed test set data to the unperturbed classifier f. Having comparable performance is understandable as 80% of the training data for f20rotation were still unperturbed. Likewise, f20rotation performed better than f no matter how much test data were perturbed (as long as some of them were perturbed). Additionally, and perhaps more importantly, the error rate of f20rotation did not significantly increase no matter how many of the test images were perturbed, indicating that this classifier performed well and consistent with its performance. So, while f may only be robust to data with a few images perturbed by rotation, perturbing the data with just 20% rotated training images appeared to create a more robust classifier than the original/given classifier. While the variances of the performance between the two classifiers across the 200 randomly perturbed test sets were comparable, neither were particularly high and, therefore, did not call the robustness of either classifier into question.
Since perturbations are random in nature, most images will, to varying degrees, be affected by multiple perturbations. To test if training with perturbed data can improve classifier robustness to simultaneous/multiple perturbations, classifiers were generated for two datasets using the original training data, and then, subsequently, a new set of perturbed classifiers was trained using training data where 10% of the training set was perturbed by each perturbation (leaving 50% unperturbed). Perturbing the training data with sets of each perturbation improved classifier robustness to multiple perturbations, as measured by the error rate, with small images and a large sample size (
Figure 7A) and larger images, but a smaller sample size (
Figure 7B). While the classifiers trained on the BUSI data showed less of a performance improvement (error rate decrease) than the PneumoniaMNIST classifiers, both still showed significant decreases in the error rate when the training data of each classifier were perturbed, as is shown in
Figure 7 and
Table A7. The smaller performance increase with the BUSI-trained classifiers may be due to the larger number of parameters, leading the models to be more sensitive to noise, and the smaller sample size of the training dataset, making it more difficult for the classifiers to learn the underlying data distribution. In addition, since there was not much similarity between the 28 × 28 chest X-ray images and 500 × 500 breast ultrasound images, it is unlikely that the classifiers were performing better due to overfitting or some dataset-specific phenomenon. Despite these differences, this approach improved the performance of the classifiers for both datasets with different sizes and applications. This implies that this approach can improve the robustness of other, similar medical imaging datasets to common perturbations. Essentially, if a set of perturbations is known to possibly affect an image, a small portion of training set images should be transformed using those perturbations. Nevertheless, these results show that training classifiers with perturbed data from several individual perturbations seems to make the classifiers more robust even to multiple simultaneous perturbations.
To put our findings into a broader context, comparisons with adversarial training were also performed. Adversarial training approaches have notable strengths, as they are the de facto best method for complex adversaries that may not be visually perceptible. They have also been shown to improve the robustness of classifiers in many different applications. Additionally, they have wider applicability by being able to be used in models like generative adversarial networks for applications such as image denoising. However, the general weaknesses of adversarial classifiers have been shown to not be able to generalize well, struggle with multiple sources of adversarial noise, and be computationally complex. Although SPSA is only one type of adversarial training, these broad weaknesses make it unlikely that implementing other forms of adversarial noise would improve robustness in this scenario. It is important to note that adversarial training can be limited in terms of which types of adversaries it can be applied to [
49]. For instance, the SPSA-trained classifier showed the same general trends in performance as the unperturbed classifier when applied to rotation-, tilt-, and blur-perturbed test data. This indicates a lack of robustness to these perturbations, which is expected since rotation, tilt, and blur affect image data differently than SPSA. Moreover, previous studies have shown that perturbing data with certain types of untargeted noise can create similar changes in performance to adversarial training [
50]. There is also not much variance in the computational complexity of each perturbation, whereas the computational requirement of adversarial training can vary significantly. More details on the computational complexity of this work is contained in
Table A10 and
Table A11. It was also demonstrated in this study that perturbing the training data can result in classifiers being robust to multiple perturbations, a notable area that adversarial models are weak in. While adversarial attacks can prepare classifiers for certain perturbations, if a specific perturbation is suspected of being possibly present in a dataset, especially if multiple perturbations are suspected, a dataset should be trained with a small number of images augmented using that perturbation.
As the previous results have shown that the approach in this study could evaluate and improve the robustness of a series of deep classifiers, future implementations of this work should continue to vary the amount of perturbed training samples to develop a robust classifier. Previous research has established the idea that classifiers robust to a perturbation should perform similarly to classifiers trained with a small amount of that perturbation [
51]. To find this small amount, it is important to thoroughly vary the amount of perturbed images in the training and test set, as was performed in this study. While the experiments in this study were performed on simulated perturbations, real-world diagnostic workflows could prepare their classifiers for noise by preemptively diversifying their training set with expected perturbations. Although this cannot circumvent the regulatory need for clinical validation tests, this approach can indicate which diagnostic classifiers are likely to be robust and, also, a liberal estimation of how robust the classifier can be.
Limitations
There are several limitations to this work that should be investigated in future studies. First, the severity of each of the perturbations was fixed for all applications in this study. In real-world diagnostic applications, noise may not be present in images at the same severity. While the SSIM data in
Table A8 imply that perturbation intensity is not directly related to robustness, future studies should investigate how robust classifiers are when the intensity/severity of each individual perturbation is varied.
Another limitation of this study is that it is unknown if this robustness will occur for all types of deep learning classifiers. For example, it is difficult to interpret exactly why a classifier is better at predicting certain images than others, as most neural networks have many trainable weights that must be accounted for. Additionally, although this study showed robustness for networks trained on grayscale images, other image modalities produce three-channel (RGB) or three-dimensional images that may require more complex classifiers. As such, future studies should investigate the impact of classifier complexity on its robustness.
One final consideration is that, while this procedure can improve the robustness of classifiers in certain scenarios, it will not necessarily create a clinic-ready classifier. Just as the adversarially trained classifier did not perform well on the images augmented using the perturbations in the study, these perturbations are not the only sources of noise an image may be subjected to. Testing a classifier under more diverse perturbations would require more iterations and, therefore, be much more computationally expensive and infeasible for most clinical applications. Furthermore, the clinical application of the classifiers involved in this study cannot be ensured without actual validation on new clinical data, which should be performed in future studies.