Novel CaLB-like Lipase Found Using ProspectBIO, a Software for Genome-Based Bioprospection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatics
2.2. Experimental
3. Results
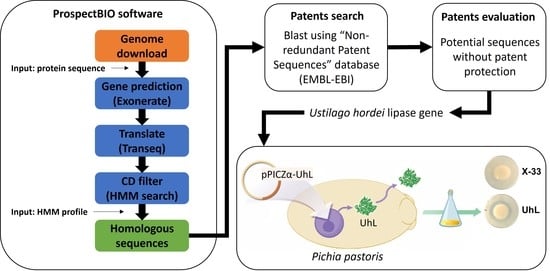
3.1. Bioprospecting
3.1.1. ProspectBIO Software
3.1.2. Prospecting a Novel CaLB-like Lipase
3.1.3. UhL Three-Dimensional Modeling
3.2. Experimental Validation
3.2.1. UhL Expression in Pichia pastoris
3.2.2. UhL Characteristics
3.2.3. Substrate Specificity
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Timson, D.J. Four Challenges for Better Biocatalysts. Fermentation 2019, 5, 39. [Google Scholar] [CrossRef] [Green Version]
- Kamble, A.; Srinivasan, S.; Singh, H. In-Silico Bioprospecting: Finding Better Enzymes. Mol. Biotechnol. 2019, 61, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Martínez-Abarca, F.; Golyshin, P.N. Mining Genomes and “metagenomes” for Novel Catalysts. Curr. Opin. Biotechnol. 2005, 16, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.E.; Boone, B.E. Next-Generation Sequencing Strategies. Cold Spring Harb. Perspect. Med. 2019, 9, a025791. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roumpeka, D.D.; Wallace, R.J.; Escalettes, F.; Fotheringham, I.; Watson, M. A Review of Bioinformatics Tools for Bio-Prospecting from Metagenomic Sequence Data. Front. Genet. 2017, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Scalzitti, N.; Jeannin-Girardon, A.; Collet, P.; Poch, O.; Thompson, J.D. A Benchmark Study of Ab Initio Gene Prediction Methods in Diverse Eukaryotic Organisms. BMC Genom. 2020, 21, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Sarmah, N.; Rani, K.Y. Recent Advances on Sources and Industrial Applications of Lipases. Biotechnol. Prog. 2018, 34, 5–28. [Google Scholar] [CrossRef]
- de Godoy Daiha, K.; Angeli, R.; de Oliveira, S.D.; Almeida, R.V. Are Lipases Still Important Biocatalysts? A Study of Scientific Publications and Patents for Technological Forecasting. PLoS ONE 2015, 10, e0131624. [Google Scholar]
- Patkar, S.; Halkier, T. Two Lipases from Candida Antarctica: Cloning and Expression in Aspergillus Oryzae. Can. J. Bot. 1995, 73, 869–875. [Google Scholar]
- Anderson, E.M.; Larsson, K.M.; Rk, O.L.E.K. One Biocatalyst Many Applications: The Use of Candida Antarctica B-Lipase in Organic Synthesis. Biocatal. Biotransformation 1998, 16, 181–204. [Google Scholar] [CrossRef]
- Uppenberg, J.; Hansen, M.T.; Patkar, S.; Jones, T.A. The Sequence, Crystal Structure Determination and Refinement of Two Crystal Forms of Lipase B from Candida Antarctica. Structure 1994, 2, 293–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senanayake, S.N.; Shahidi, F. Lipase-Catalyzed Incorporation of Docosahexaenoic Acid (DHA) into Borage Oil: Optimization Using Response Surface Methodology. Food Chem. 2002, 77, 115–123. [Google Scholar] [CrossRef]
- Baldessari, A.; Iglesias, L.E. Lipases in Green Chemistry: Acylation and Alcoholysis on Steroids and Nucleosides. In Lipases and Phospholipases; Humana Press: Totowa, NJ, USA, 2012; Volume 861, pp. 457–469. [Google Scholar]
- Gog, A.; Roman, M.; Toşa, M.; Paizs, C.; Irimie, F.D. Biodiesel Production Using Enzymatic Transesterification-Current State and Perspectives. Renew. Energy 2012, 39, 10–16. [Google Scholar] [CrossRef]
- Kapoor, M.; Gupta, M.N. Lipase Promiscuity and Its Biochemical Applications. Process Biochem. 2012, 47, 555–569. [Google Scholar] [CrossRef]
- de Miranda, A.S.; Miranda, L.S.M.; de Souza, R.O.M.A. Lipases: Valuable Catalysts for Dynamic Kinetic Resolutions. Biotechnol. Adv. 2015, 33, 372–393. [Google Scholar] [CrossRef]
- Robert, J.M.; Lattari, F.S.; Machado, A.C.; de Castro, A.M.; Almeida, R.V.; Torres, F.A.G.; Valero, F.; Freire, D.M.G. Production of Recombinant Lipase B from Candida Antarctica in Pichia Pastoris under Control of the Promoter PGK Using Crude Glycerol from Biodiesel Production as Carbon Source. Biochem. Eng. J. 2017, 118, 123–131. [Google Scholar] [CrossRef]
- Robert, J.M.; Betancur, M.O.; Carlos, A.; Machado, O.; Arruda, A.; Castelo, V.; Reis, B.; Almeida, R.V.; Araripe, F.; Torres, G.; et al. Increase of Candida Antarctica Lipase B Production under PGK Promoter in Pichia Pastoris: Effect of Multicopies. Braz. J. Microbiol. 2019, 50, 405–413. [Google Scholar] [CrossRef] [PubMed]
- de Macedo Robert, J.; Garcia-Ortega, X.; Montesinos-Seguí, J.L.; Guimaraes Freire, D.M.; Valero, F. Continuous Operation, a Realistic Alternative to Fed-Batch Fermentation for the Production of Recombinant Lipase B from Candida Antarctica under the Constitutive Promoter PGK in Pichia Pastoris. Biochem. Eng. J. 2019, 147, 39–47. [Google Scholar] [CrossRef]
- Tang, S.; Boehme, L.; Lam, H.; Zhang, Z. Pichia Pastoris Fermentation for Phytase Production Using Crude Glycerol from Biodiesel Production as the Sole Carbon Source. Biochem. Eng. J. 2009, 43, 157–162. [Google Scholar] [CrossRef]
- Buerth, C.; Kovacic, F.; Stock, J.; Terfrüchte, M.; Wilhelm, S.; Jaeger, K.; Feldbrügge, M.; Schipper, K.; Ernst, J.F.; Tielker, D. Uml2 Is a Novel CalB-Type Lipase of Ustilago Maydis with Phospholipase A Activity. Appl. Microbiol. Biotechnol. 2014, 98, 4963–4973. [Google Scholar] [CrossRef]
- Vaquero, M.E.; De Eugenio, L.I.; Martínez, M.J.; Barriuso, J. A Novel CalB-Type Lipase Discovered by Fungal Genomes Mining. PLoS ONE 2015, 10, e0124882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Widmann, M.; Juhl, P.B.; Pleiss, J. Structural Classification by the Lipase Engineering Database: A Case Study of Candida Antarctica Lipase A. BMC Genom. 2010, 11, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; McWilliam, H.; de la Torre, A.R.; Grodowski, A.; Benediktovich, I.; Goujon, M.; Nauche, S.; Lopez, R. Non-Redundant Patent Sequence Databases with Value-Added Annotations at Two Levels. Nucleic Acids Res. 2009, 38, D52–D56. [Google Scholar] [CrossRef] [Green Version]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of Protein Structures: Patterns of Nonbonded Atomic Interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Cregg, J.M.; Russell, K.A. Transformation. In Pichia Protocols. Methods in Molecular Biology; Higgins, D.R., Cregg, J.M., Eds.; Humana Press: Berlin, Germany, 1998; Volume 103, pp. 27–39. [Google Scholar]
- Lõoke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of Genomic DNA from Yeasts for PCR-Based Applications. Biotechniques 2011, 50, 325–328. [Google Scholar] [CrossRef]
- Prim, N.; Sánchez, M.; Ruiz, C.; Pastor, F.I.J.; Diaz, P. Use of Methylumbeliferyl-Derivative Substrates for Lipase Activity Characterization. J. Mol. Catal. B Enzym. 2003, 22, 339–346. [Google Scholar] [CrossRef]
- Slater, G.S.C.; Birney, E. Automated Generation of Heuristics for Biological Sequence Comparison. BMC Bioinform. 2005, 6, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pleiss, J.; Fischer, M.; Peiker, M.; Thiele, C.; Schmid, R.D. Lipase Engineering Database Understanding and Exploiting Sequence–Structure–Function Relationships. J. Mol. Catal. B Enzym. 2000, 10, 491–508. [Google Scholar] [CrossRef]
- Kugimiya, W.; Otani, Y.; Hashimoto, Y.; Takagi, Y. Molecular Cloning and Nucleotide Sequence of the Lipase Gene from Pseudomonas Fragi. Biochem. Biophys. Res. Commun. 1986, 141, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Classen, T.; Kovacic, F.; Lauinger, B.; Pietruszka, J.; Jaeger, K. Screening for Enantioselective Lipases. In Hydrocarbon and Lipid Microbiology Protocols; Springer: Berlin/Heidelberg, Germany, 2016; pp. 37–69. [Google Scholar]
- Eom, G.T.; Lee, S.H.; Song, B.K.; Chung, K.; Kim, Y.; Song, J.K. High-Level Extracellular Production and Characterization of Candida Antarctica Lipase B in Pichia Pastoris. J. Biosci. Bioeng. 2013, 116, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.T.N.; Fojan, P.; Petersen, S.B. How Do Lipases and Esterases Work: The Electrostatic Contribution. J. Biotechnol. 2001, 85, 115–147. [Google Scholar] [CrossRef]
- Blank, K.; Morfill, J.; Gumpp, H.; Gaub, H.E. Functional Expression of Candida Antarctica Lipase B in Eschericha coli. J. Biotechnol. 2006, 125, 474–483. [Google Scholar] [CrossRef]
- Wittrup, M.; Bornscheuer, U.T.; Hult, K. Expression of Candida Antarctica Lipase B in Pichia Pastoris and Various Escherichia coli Systems. Protein Expr. Purif. 2008, 62, 90–97. [Google Scholar]
- Benkert, P.; Tosatto, S.C.E.; Schomburg, D. QMEAN: A Comprehensive Scoring Function for Model Quality Assessment. Proteins Struct. Funct. Bioinform. 2008, 71, 261–277. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-dannert, C. Recombinant Microbial Lipases for Biotechnological Applications. Bioorg. Med. Chem. 1999, 7, 2123–2130d. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Anumanthan, A.; Gao, X.; Ilangovan, K.; Suzara, V.V.; Düzgüneş, N.; Renugopalakrishnan, V. Expression of Recombinant Proteins in Pichia Pastoris. Appl. Biochem. Biotechnol. 2007, 142, 105–124. [Google Scholar] [CrossRef]
- Schein, C.H.; Noteborn, M.H.M. Formation Od Soluble Recombinant Proteins on Escherichia Coli Is Favored by Lower Growth Temperature. Nat. Biotechnol. 1988, 6, 291–294. [Google Scholar] [CrossRef]
- Dragosits, M.; Stadlmann, J.; Albiol, J.; Baumann, K.; Maurer, M.; Gasser, B.; Sauer, M.; Altmann, F.; Ferrer, P.; Mattanovich, D.; et al. The Effect of Temperature on the Proteome of Recombinant Pichia Pastoris Research Articles. J. Proteome Res. 2009, 8, 1380–1392. [Google Scholar] [CrossRef]
- Jahic, M.; Gusta, M.; Jansen, A.; Martinelle, M.; Enfors, S. Analysis and Control of Proteolysis of a Fusion Protein in Pichia Pastoris Fed-Batch Processes. J. Biotechnol. 2003, 102, 3–8. [Google Scholar] [CrossRef]
- Jahic, M.; Wallberg, F.; Bollok, M.; Garcia, P.; Enfors, S. Temperature Limited Fed-Batch Technique for Control of Proteolysis in Pichia Pastoris Bioreactor Cultures. Microb. Cell Fact. 2003, 2, 6. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, Z.; Xu, Q.; Du, G.; Hua, Z.; Liu, L.; Li, J.; Chen, J. Lowering Induction Temperature for Enhanced Production of Polygalacturonate Lyase in Recombinant Pichia Pastoris. Process Biochem. 2009, 44, 949–954. [Google Scholar] [CrossRef]
- Wan, L.; Cai, H.; Yang, H.; Lu, Y.; Li, Y.; Li, X.; Li, S.; Zhang, J.; Li, Y.; Cheng, J.; et al. High-Level Expression of a Functional Humanized Single-Chain Variable Fragment Antibody against CD25 in Pichia Pastoris. Appl. Microbiol. Biotechnol. 2008, 81, 33–41. [Google Scholar] [CrossRef]
- Duan, H.; Wang, H.; Ma, B.; Jiang, P.; Tu, P.; Ni, Z.; Li, X.; Li, M.; Ma, X.; Wang, B.; et al. Codon Optimization and Expression of Irisin in Pichia Pastoris GS115. Int. J. Biol. Macromol. 2015, 79, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Meinander, N.Q.; Jönsson, L.J. Fermentation Strategies for Improved Heterologous Expression of Laccase in Pichia Pastoris. Biotechnol. Bioeng. 2002, 79, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Invitrogen-ThermoFisher. Pichia Expression Kit: For Expression of Recombinant Proteins in Pichia pastoris. Available online: https://tools.thermofisher.com/content/sfs/manuals/pich_man.pdf (accessed on 1 January 2023).
- Stratton, J.; Chiruvolu, V.; Meagher, M. High Cell-Density Fermentation. Pichia Protoc. Hum. Press 1997, 103, 107–120. [Google Scholar]
- Minning, S.; Serrano, A.; Ferrer, P.; Sola, C.; Schmid, R.D.; Valero, F. Optimization of the High-Level Production of Rhizopus Oryzae Lipase in Pichia Pastoris. J. Biotechnol. 2001, 86, 59–70. [Google Scholar] [CrossRef] [PubMed]





| Accession Numbers | Organism | Protein Length | ID% |
|---|---|---|---|
| ACI06118 | |||
| 1TCA | Candida antarctica | 342 | 100 |
| P41365 | |||
| CCF54401 | Ustilago hordei | 343 | 76 |
| CP010917 | Sporisorium scitamineum SSC39 | 339 | 74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brêda, G.C.; Faria, P.E.; Rodrigues, Y.S.; Pinheiro, P.B.; Nucci, M.C.R.; Ferrer, P.; Freire, D.M.G.; Almeida, R.V.; Mesquita, R.D. Novel CaLB-like Lipase Found Using ProspectBIO, a Software for Genome-Based Bioprospection. BioTech 2023, 12, 6. https://doi.org/10.3390/biotech12010006
Brêda GC, Faria PE, Rodrigues YS, Pinheiro PB, Nucci MCR, Ferrer P, Freire DMG, Almeida RV, Mesquita RD. Novel CaLB-like Lipase Found Using ProspectBIO, a Software for Genome-Based Bioprospection. BioTech. 2023; 12(1):6. https://doi.org/10.3390/biotech12010006
Chicago/Turabian StyleBrêda, Gabriela C., Priscila E. Faria, Yuri S. Rodrigues, Priscila B. Pinheiro, Maria Clara R. Nucci, Pau Ferrer, Denise M. G. Freire, Rodrigo V. Almeida, and Rafael D. Mesquita. 2023. "Novel CaLB-like Lipase Found Using ProspectBIO, a Software for Genome-Based Bioprospection" BioTech 12, no. 1: 6. https://doi.org/10.3390/biotech12010006