Relevant Aspects in the Development of Electrochemical Aptasensors for the Determination of Antibiotics—A Review
Abstract
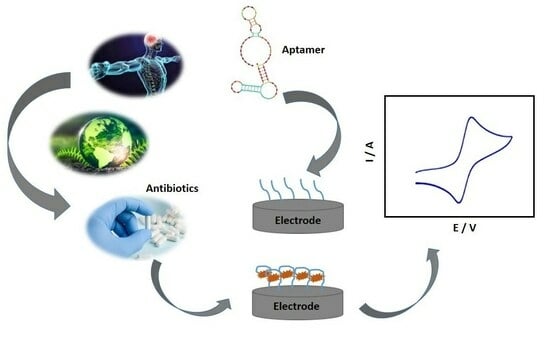
:1. Introduction
2. Antibiotics and Electrochemical Detection
3. Relevant Aspects in the Development of Electrochemical Aptasensors
3.1. Nanomaterial-Modified Electrochemical Aptasensors
3.2. Configurations of Electrochemical Aptasensors
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fang, P.; Qu, H.; Mao, Y.; Zheng, L. Aptamers for mycotoxin recognition in food: Recent advances and future considerations. Adv. Agrochem 2023, 2, 213–220. [Google Scholar] [CrossRef]
- Uğurlu, O.; Man, E.; Gök, O.; Ülker, G.; Soytürk, H.; Özyurt, C.; Evran, S. A review of aptamer-conjugated nanomaterials for analytical sample preparation: Classification according to the utilized nanomaterials. Anal. Chim. Acta 2023, 342001. [Google Scholar] [CrossRef]
- Di Mauro, V.; Lauta, F.C.; Modica, J.; Appleton, S.L.; De Franciscis, V.; Catalucci, D. Diagnostic and Therapeutic Aptamers: A Promising Pathway to Improved Cardiovascular Disease Management. JACC Basic Transl. Sci. 2023. [Google Scholar] [CrossRef]
- Wang, L.; Peng, X.; Fu, H.; Huang, C.; Li, Y.; Liu, Z. Recent advances in the development of electrochemical aptasensors for detection of heavy metals in food. Biosens. Bioelectron. 2020, 147, 111777. [Google Scholar] [CrossRef] [PubMed]
- Torres-Vázquez, B.; de Lucas, A.M.; García-Crespo, C.; García-Martín, J.A.; Fragoso, A.; Fernández-Algar, M.; Perales, C.; Domingo, E.; Moreno, M.; Briones, C. In vitro Selection of High Affinity DNA and RNA Aptamers that Detect Hepatitis C Virus Core Protein of Genotypes 1 to 4 and Inhibit Virus Production in Cell Culture. J. Mol. Biol. 2022, 434, 167501. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Hussain, A.; Fahimi, H.; Aliakbari, F.; Bloukh, S.H.; Edis, Z.; Babadaei, M.M.N.; Izadi, Z.; Varnamkhasti, B.S.; Jahanshahi, F.; et al. A review on the therapeutic applications of aptamers and aptamer-conjugated nanoparticles in cancer, inflammatory and viral diseases. Arab. J. Chem. 2022, 15, 103626. [Google Scholar] [CrossRef]
- Nur, Y.; Gaffar, S.; Hartati, Y.W.; Subroto, T. Applications of electrochemical biosensor of aptamers-based (APTASENSOR) for the detection of leukemia biomarker. Sens. Bio-Sens. Res. 2021, 32, 100416. [Google Scholar] [CrossRef]
- Yu, H.; Alkhamis, O.; Canoura, J.; Liu, Y.; Xiao, Y. Advances and Challenges in Small-Molecule DNA Aptamer Isolation. Charact. Sens. Dev. Angew. Chem. Int. Ed. Engl. 2021, 60, 16800–16823. [Google Scholar] [CrossRef] [PubMed]
- Alkhamis, O.; Canoura, J.; Yu, H.; Liu, Y.; Xiao, Y. Innovative engineering and sensing strategies for aptamer-based small-molecule detection. TrAC Trends Anal. Chem. 2019, 121, 115699. [Google Scholar] [CrossRef]
- Lin, X.; Sun, X.; Luo, S.; Liu, B.; Yang, C. Development of DNA-based signal amplification and microfluidic technology for protein assay: A review. TrAC Trends Anal. Chem. 2016, 80, 132–148. [Google Scholar] [CrossRef]
- Xie, Y.; Huang, Y.; Li, J.; Wu, J. A trigger-based aggregation of aptamer-functionalized gold nanoparticles for colorimetry An example on detection of Escherichia coli O157:H7. Sens. Actuators B Chem. 2021, 339, 129865. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Q. Competitive fluorescence assay for Cd2+ based on aptamer structure-switching. Microchem. J. 2023, 194, 109348. [Google Scholar] [CrossRef]
- Yang, J.; Liu, H.; Huang, Y.; Li, L.; Zhu, X.; Ding, Y. One-step hydrothermal synthesis of near-infrared emission carbon quantum dots as fluorescence aptamer sensor for cortisol sensing and imaging. Talanta 2023, 260, 124637. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Back, H.; Joo, H.J.; Lim, D.-S.; Lee, J.E.; Lee, H.J. Simultaneous detection method for two cardiac disease protein biomarkers on a single chip modified with mixed aptamers using surface plasmon resonance. Talanta 2024, 267, 125232. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Prakash, N.; Paul, D.; Mukherji, S. Anti-nucleolin aptamer mediated specific detection of cancer cells by Localized Surface Plasmon Resonance-based U-bent optical fiber. Biosens. Bioelectron. X 2023, 13, 100318. [Google Scholar] [CrossRef]
- Zhao, S.; Huang, J.; Li, D.; Yang, L. Aptamer-based chemiluminescent optical fiber immunosensor with enhanced signal amplification for ultrasensitive detection of tumor biomarkers. Biosens. Bioelectron. 2022, 214, 114505. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Li, J.; Wu, Y.; Cao, X.; Zhang, Z. Rational design of hairpin aptamer using intrinsic disorder mechanism to enhance sensitivity of aptamer folding-based electrochemical sensor for tobramycin. Sens. Actuators B Chem. 2023, 394, 134354. [Google Scholar] [CrossRef]
- Kim, E.R.; Dang, T.T.-T.; Lee, S.J.; Nguyen, T.T.-Q.; Park, J.-W.; Gu, M.B. A highly sensitive sandwich-type electrochemical aptasensor using a pair of novel truncated aptamers for the detection of vaspin. Chem. Eng. J. 2023, 477, 147002. [Google Scholar] [CrossRef]
- Khan, R.; Deshpande, A.S.; Proteasa, G.; Andreescu, S. Aptamer-based electrochemical biosensor with S protein binding affinity for COVID-19 detection: Integrating computational design with experimental validation of S protein binding affinity. Sens. Actuators B Chem. 2024, 399, 134775. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, W.; Lv, C.; Liu, X.; Yang, M.; Guo, M.; Fu, Q. Electrochemical biosensors represent promising detection tools in medical field. Adv. Sens. Energy Mater. 2023, 2, 100081. [Google Scholar] [CrossRef]
- Zhong, S.; Chen, L.; Shi, X.; Chen, G.; Sun, D.; Zhang, L. Recent advances in electrochemical aptasensors for detecting cardiac biomarkers: A review. Microchem. J. 2023, 193, 109063. [Google Scholar] [CrossRef]
- Azzouz, A.; Hejji, L.; Kumar, V.; Kim, K.-H. Nanomaterials-based aptasensors: An efficient detection tool for heavy-metal and metalloid ions in environmental and biological samples. Environ. Res. 2023, 238 Pt 1, 117170. [Google Scholar] [CrossRef] [PubMed]
- Ikebukuro, K.; Kiyohara, C.; Sode, K. Electrochemical detection of protein using a double aptamer sandwich. Anal. Lett. 2004, 37, 2901–2909. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, S.; Ma, J.; Zhou, X.; Sun, X.; Jing, H.; Lin, M.; Zhou, C. Enzyme-catalyzed electrochemical aptasensor for ultrasensitive detection of soluble PD-L1 in breast cancer based on decorated covalent organic frameworks and carbon nanotubes. Anal. Chim. Acta 2023, 1282, 341927. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, H.; Li, X.; Huang, H.; Bian, H.; Liang, J.; Zhou, Z. A label-free electrochemical aptasensor for low-density lipoprotein detection using MoS2-Au-Fc nanosheets as a high-performance redox indicator. Microchem. J. 2023, 193, 109068. [Google Scholar] [CrossRef]
- Chen, K.; Zhao, H.; Wang, Z.; Zhou, F.; Lan, M. Sandwich-type electrochemical aptasensor based on polydopamine-Au-metal ions as signal label and coralloid Au-conductive carbon architecture-modified electrode for the simultaneous detection of cardiac troponin I and myoglobin. Sens. Actuators B Chem. 2023, 390, 134044. [Google Scholar] [CrossRef]
- Chen, Y.; Gu, Y.; Yi, X.; Huang, H.; Li, Y.; Yang, B.; Guo, S.; Bai, L. Conductive nano-gold self-assembled MXene@hemin with high catalytic activity achieved by strong metal-support interactions: A powerful nanozyme for development of electrochemical aptasensor in tuberculosis diagnosis. Chem. Eng. J. 2023, 466, 143112. [Google Scholar] [CrossRef]
- Li, L.; Ma, R.; Wang, W.; Zhang, L.; Li, J.; Eltzov, E.; Wang, S.; Mao, X. Group-targeting aptamers and aptasensors for simultaneous identification of multiple targets in foods. TrAC Trends Anal. Chem. 2023, 166, 117169. [Google Scholar] [CrossRef]
- Melinte, G.; Hosu, O.; Cristea, C.; Marrazza, G. Ara H1 peanut allergen detection using a labelled electrochemical aptasensor based on GO-COOH@bimetallic composite platform. Food Chem. 2023, 400, 134074. [Google Scholar] [CrossRef]
- Kang, M.; Yao, Y.; Yuan, B.; Zhang, S.; Oderinde, O.; Zhang, Z. A sensitive bimetallic copper/bismuth metal-organic frameworks-based aptasensors for zearalenone detection in foodstuffs. Food Chem. 2024, 437 Pt 1, 137827. [Google Scholar] [CrossRef]
- Lai, Z.; Mahdavi, B.; Baghayeri, M. Label-free and sensitive determination of toxic Cd(II) in environmental waters using a Fe3O4-PEI-Au based electrochemical aptasensor. Alex. Eng. J. 2023, 83, 251–256. [Google Scholar] [CrossRef]
- Zhang, Z.; Karimi-Maleh, H. In situ synthesis of label-free electrochemical aptasensor-based sandwich-like AuNPs/PPy/Ti3C2Tx for ultrasensitive detection of lead ions as hazardous pollutants in environmental fluids. Chemosphere 2023, 324, 138302. [Google Scholar] [CrossRef] [PubMed]
- Yalagandula, B.P.; Mohanty, S.; Goswami, P.P.; Singh, S.G. Optimizations towards a nearly invariable WO3-functionalized electrochemical aptasensor for ultra-trace identification of arsenic in lake water. Sens. Actuators B Chem. 2024, 398, 134730. [Google Scholar] [CrossRef]
- Khosropour, H.; Keramat, M.; Laiwattanapaisal, W. A dual action electrochemical molecularly imprinted aptasensor for ultra-trace detection of carbendazim. Biosens. Bioelectron. 2024, 243, 115754. [Google Scholar] [CrossRef] [PubMed]
- Zein, M.I.H.L.; Hardianto, A.; Irkham, I.; Zakiyyah, S.N.; Devi, M.J.; Manan, N.S.A.; Ibrahim, A.U.; Hartati, Y.W. Recent development of electrochemical and optical aptasensors for detection of antibiotics in food monitoring applications. J. Food Compos. Anal. 2023, 124, 105644. [Google Scholar] [CrossRef]
- Joshi, A.; Kim, K.-H. Recent advances in nanomaterial-based electrochemical detection of antibiotics: Challenges and future perspectives. Biosens. Bioelectron. 2020, 153, 112046. [Google Scholar] [CrossRef] [PubMed]
- Hui, X.; Fang, W.; Wang, G.; Liu, H.; Dai, X. Waste recycling of antibiotic mycelial residue: The feasible harmless treatment and source control of antibiotic resistance. J. Clean. Prod. 2023, 401, 136786. [Google Scholar] [CrossRef]
- Ding, R.; Chen, Y.; Wang, Q.; Wu, Z.; Zhang, X.; Li, B.; Lin, L. Recent advances in quantum dots-based biosensors for antibiotics detection. J. Pharm. Anal. 2022, 12, 355–364. [Google Scholar] [CrossRef]
- Yu, C.; Pang, H.; Wang, J.-H.; Chi, Z.-Y.; Zhang, Q.; Kong, F.-T.; Xu, Y.-P.; Li, S.-Y.; Che, J. Occurrence of antibiotics in waters, removal by microalgae-based systems, and their toxicological effects: A review. Sci. Total Environ. 2022, 813, 151891. [Google Scholar] [CrossRef]
- Singh, H.; Thakur, B.; Bhardwaj, S.K.; Khatri, M.; Kim, K.-H.; Bhardwaj, N. Nanomaterial-based fluorescent biosensors for the detection of antibiotics in foodstuffs: A review. Food Chem. 2023, 426, 136657. [Google Scholar] [CrossRef]
- Wang, Q.; Zhao, W.-M. Optical methods of antibiotic residues detections: A comprehensive review. Sens. Actuators B Chem. 2018, 269, 238–256. [Google Scholar] [CrossRef]
- Fu, L.; Mao, S.; Chen, F.; Zhao, S.; Su, W.; Lai, G.; Yu, A.; Lin, C.-T. Graphene-based electrochemical sensors for antibiotic detection in water, food and soil: A scientometric analysis in CiteSpace (2011–2021). Chemosphere 2022, 297, 134127. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Wei, Y.; Wang, J.; Yu, Y. A photo-renewable ZIF-8 photo electrochemical sensor for the sensitive detection of sulfamethoxazole antibiotic. Anal. Chim. Acta 2021, 1178, 338793. [Google Scholar] [CrossRef] [PubMed]
- Sgobbi, L.F.; Razzino, C.A.; Machado, S.A.S. A disposable electrochemical sensor for simultaneous detection of sulfamethoxazole and trimethoprim antibiotics in urine based on multiwalled nanotubes decorated with Prussian blue nanocubes modified screen-printed electrode. Electrochim. Acta 2016, 191, 1010–1017. [Google Scholar] [CrossRef]
- Elfiky, M.; Salahuddin, N.; Hassanein, A.; Matsuda, A.; Hattori, T. Detection of antibiotic Ofloxacin drug in urine using electrochemical sensor based on synergistic effect of different morphological carbon materials. Microchem. J. 2019, 146, 170–177. [Google Scholar] [CrossRef]
- Bai, Z.; Chen, Y.; Li, F.; Zhou, Y.; Yin, H.; Ai, S. Electrochemical aptasensor for sulfadimethoxine detection based on the triggered cleavage activity of nuclease P1 by aptamer-target complex. Talanta 2019, 204, 409–414. [Google Scholar] [CrossRef]
- Li, F.; Wu, Y.; Chen, D.; Guo, Y.; Wang, X.; Sun, X. Sensitive dual-labeled electrochemical aptasensor for simultaneous detection of multi-antibiotics in milk. Int. J. Hydrogen Energy 2021, 46, 23301–23309. [Google Scholar] [CrossRef]
- Guan, J.; He, K.; Gunasekaran, S. Selection of ssDNA aptamer using GO-SELEX and development of DNA nanostructure-based electrochemical aptasensor for penicillin. Biosens. Bioelectron. X 2022, 12, 100220. [Google Scholar] [CrossRef]
- Naseri, M.; Niazi, A.; Bagherzadeh, K.; Konoz, E.; Samadikhah, H.R. Modified electrochemical aptasensor for ultrasensitive detection of tetracycline: In silico and in vitro studies. Food Chem. 2023, 421, 136195. [Google Scholar] [CrossRef]
- Wei, X.; Yin, M.; Zhang, L.; Sun, Y.; Luo, Y.; Xu, D. Octahedral Cu2O nanomaterials as electrochemical aptasensor for sensitive detection of tetracycline in milk. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2024, 304, 123361. [Google Scholar] [CrossRef]
- Vanani, S.M.; Izadi, Z.; Hemmati, R.; Saffar, B. Fabrication of an ultrasensitive aptasensor for precise electrochemical detection of the trace amounts of streptomycin in milk. Colloids Surf. B Biointerfaces 2021, 206, 111964. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yan, W.; Guo, Y.; Wang, X.; Zhang, F.; Yu, L.; Guo, C.; Fang, G. Sensitive and selective electrochemical aptasensor via diazonium-coupling reaction for label-free determination of oxytetracycline in milk samples. Sens. Actuators Rep. 2020, 2, 100009. [Google Scholar] [CrossRef]
- Li, F.; Zhu, J.; Liu, Y.; Lil, Z.; Kang, H.; Li, R. Magnetic bead-based electrochemical aptasensor doped with multi-wall carbon nanotubes for the detection of ampicillin in milk. Int. J. Electrochem. Sci. 2020, 15, 7520–7530. [Google Scholar] [CrossRef]
- Song, J.; Huang, M.; Jiang, N.; Zheng, S.; Mu, T.; Meng, L.; Liu, Y.; Liu, J.; Chen, G. Ultrasensitive detection of amoxicillin by TiO2-g-C3N4@AuNPs impedimetric aptasensor: Fabrication, optimization, and mechanism. J. Hazard. Mater. 2020, 391, 122024. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yin, H.; Zhang, Y.; Wang, L.; Du, Y.; Zhuge, Y.; Ai, S. Electrochemical aptasensor for ampicillin detection based on the protective effect of aptamer-antibiotic conjugate towards DpnII and Exo III digestion. Talanta 2019, 197, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hu, M.; Wang, M.; Song, Y.; Zhou, N.; He, L.; Zhang, Z. Novel nanoarchitecture of Co-MOF-on-TPN-COF hybrid: Ultralowly sensitive bioplatform of electrochemical aptasensor toward ampicillin. Biosens. Bioelectron. 2019, 123, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yan, X.; Zhao, L.; Qi, X.; Wang, S.; Liang, X. An aptamer cocktail-based electrochemical aptasensor for direct capture and rapid detection of tetracycline in honey. Microchem. J. 2019, 150, 104179. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, X.; Wu, M.; Shi, M.; Tian, Q.; Fu, L.; Tsai, H.-S.; Xie, W.-F.; Lai, G.; Wang, G.; et al. A novel electrochemical aptasensor based on eco-friendly synthesized titanium dioxide nanosheets and polyethyleneimine grafted reduced graphene oxide for ultrasensitive and selective detection of ciprofloxacin. Anal. Chim. Acta 2023, 1275, 341607. [Google Scholar] [CrossRef]
- Yin, J.; Guo, W.; Qin, X.; Zhao, J.; Pei, M.; Ding, F. A sensitive electrochemical aptasensor for highly specific detection of streptomycin based on the porous carbon nanorods and multifunctional graphene nanocomposites for signal amplification. Sens. Actuators B Chem. 2017, 241, 151–159. [Google Scholar] [CrossRef]
- Song, H.-Y.; Kang, T.-F.; Lu, L.-P.; Cheng, S.-Y. Highly sensitive aptasensor based on synergetic catalysis activity of MoS2-Au-HE composite using cDNA-Au-GOD for signal amplification. Talanta 2017, 164, 27–33. [Google Scholar] [CrossRef]
- Li, S.; He, B.; Liang, Y.; Wang, J.; Jiao, Q.; Liu, Y.; Guo, R.; Wei, M.; Jin, H. Sensitive electrochemical aptasensor for determination of sulfaquinoxaline based on AuPd NPs@UiO-66-NH2/CoSe2 and RecJf exonuclease-assisted signal amplification. Anal. Chim. Acta 2021, 1182, 338948. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Xu, Q.; Zhang, Q.; Guo, Y.; Sun, X.; Wang, X.Y. Study on Aptasensors Modified by Ionic Liquid-Fe3O4 Based on Microarray Electrodes for Tetracycline Detection. Int. J. Electrochem. Sci. 2016, 11, 1699–1706. [Google Scholar] [CrossRef]
- Zhan, F.; Zhao, Y.; Dai, X.; Zeng, J.; Wang, Q. Electrochemically synthesized polyanine@Cu-BTC MOF as a bifunctional matrix for aptasensing of tetracycline in aquatic products. Microchem. J. 2024, 196, 109512. [Google Scholar] [CrossRef]
- Nie, J.; Yuan, L.; Jin, K.; Han, X.; Tian, Y.; Zhou, N. Electrochemical detection of tobramycin based on enzymes-assisted dual signal amplification by using a novel truncated aptamer with high affinity. Biosens. Bioelectron. 2018, 122, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Hu, B.; Yang, C.; Zhang, Z.; He, L.; Fang, S.; Qu, X.; Zhang, Q. Electrochemical biosensing based on protein-directed carbon nanospheres embedded with SnOx and TiO2 nanocrystals for sensitive detection of tobramycin. Biosens. Bioelectron. 2018, 99, 176–185. [Google Scholar] [CrossRef]
- Li, F.; Wang, X.; Sun, X.; Guo, Y.; Zhao, W. A dual-signal amplification strategy for kanamycin based on ordered mesoporous carbon-chitosan/gold nanoparticles-streptavidin and ferrocene labelled DNA. Anal. Chim. Acta 2018, 1033, 185–192. [Google Scholar] [CrossRef]
- Aghajari, R.; Azadbakht, A. Amplified detection of streptomycin using aptamer-conjugated palladium nanoparticles decorated on chitosan-carbon nanotube. Anal. Biochem. 2018, 547, 57–65. [Google Scholar] [CrossRef]
- Yalikun, N.; Mamat, X.; Li, Y.; Hu, X.; Wågberg, T.; Dong, Y.; Hu, G. Synthesis of an iron-nitrogen co-doped ordered mesoporous carbon-silicon nanocomposite as an enhanced electrochemical sensor for sensitive and selective determination of chloramphenicol. Colloids Surf. B Biointerfaces 2018, 172, 98–104. [Google Scholar] [CrossRef]
- Yuan, R.; Yan, Z.; Shaga, A.; He, H. Design and fabrication of an electrochemical sensing platform based on a porous organic polymer for ultrasensitive ampicillin detection. Sens. Actuators B Chem. 2021, 327, 128949. [Google Scholar] [CrossRef]
- Mahmoudpour, M.; Kholafazad-kordasht, H.; Dolatabadi, J.E.N.; Hasanzadeh, M.; Rad, A.H.; Torbati, M. Sensitive aptasensing of ciprofloxacin residues in raw milk samples using reduced graphene oxide and nanogold-functionalized poly(amidoamine) dendrimer: An innovative apta-platform towards electroanalysis of antibiotics. Anal. Chim. Acta 2021, 1174, 338736. [Google Scholar] [CrossRef]
- Qin, X.; Yin, Y.; Yu, H.; Guo, W.; Pei, M. A novel signal amplification strategy of an electrochemical aptasensor for kanamycin, based on thionine functionalized graphene and hierarchical nanoporous PtCu. Biosens. Bioelectron. 2016, 77, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Jahanbani, S.; Benvidi, A. Comparison of two fabricated aptasensors based on modified carbon paste/oleic acid and magnetic bar carbon paste/Fe3O4@oleic acid nanoparticle electrodes for tetracycline detection. Biosens. Bioelectron. 2016, 85, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Lin, X.; Jiang, N.; Huang, M. Carbon-doped WO3 electrochemical aptasensor based on Box-Behnken strategy for highly-sensitive detection of tetracycline. Food Chem. 2022, 367, 130564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Lv, L.; Ma, X.; Xie, L.; Lin, M.; Chen, H.; He, B. Au@ZnNi-MOF labeled electrochemical aptasensor for detection of enrofloxacin based on AuPt@h-CeO2/MoS2 and DNAzyme-driven DNA walker triple amplification signal strategy. Biosens. Bioelectron. 2022, 210, 114296. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Dong, S.; Gai, P.; Duan, R.; Li, F. Highly sensitive homogeneous electrochemical aptasensor for antibiotic residues detection based on dual recycling amplification strategy. Biosens. Bioelectron. 2016, 82, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Gan, N.; Li, T.; Wang, Y.; Xu, Q.; Chen, Y. An electrochemical aptasensor for multiplex antibiotics detection using Y-shaped DNA-based metal ions encoded probes with NMOF substrate and CSRP target-triggered amplification strategy. Anal. Chim. Acta 2017, 968, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Yan, X.; Li, M.; Liu, H.; Li, J.; Wang, L.; Wang, K.; Lu, X.; Wang, S.; He, B. Ultrasensitive detection of chloramphenicol using electrochemical aptamer sensor: A mini review. Electrochem. Commun. 2020, 120, 106835. [Google Scholar] [CrossRef]
- Liu, Y.; Deng, Y.; Li, S.; Chow, F.W.-N.; Liu, M.; He, N. Monitoring and detection of antibiotic residues in animal derived foods: Solutions using aptamers. Trends Food Sci. Technol. 2022, 125, 200–235. [Google Scholar] [CrossRef]
- Xie, J.; Zhang, L.; Liu, Z.; Ling, G.; Zhang, P. Application of electrochemical sensors based on nanomaterials modifiers in the determination of antipsychotics. Colloids Surf. B Biointerfaces 2022, 214, 112442. [Google Scholar] [CrossRef]
- Kaya, S.I.; Yıldırım, S.; Cetinkaya, A.; Erkmen, C.; Uslu, B.; Ozkan, S.A. Nanomaterial-based electroanalytical sensors for the selected prohibited anabolic agents, hormones and metabolic modulators and their sensitive assays. TrAC Trends Anal. Chem. 2021, 145, 116457. [Google Scholar] [CrossRef]
- Parihar, A.; Choudhary, N.K.; Sharma, P.; Khan, R. Carbon nanomaterials-based electrochemical aptasensor for point-of-care diagnostics of cancer biomarkers. Mater. Today Chem. 2023, 30, 101499. [Google Scholar] [CrossRef]
- Gañán, J.; Martínez-García, G.; Morante-Zarcero, S.; Pérez-Quintanilla, D.; Sierra, I. Nanomaterials-modified electrochemical sensors for sensitive determination of alkaloids: Recent trends in the application to biological, pharmaceutical and agri-food samples. Microchem. J. 2023, 184 Pt A, 108136. [Google Scholar] [CrossRef]
- Hui, Y.; Yang, D.; Wang, W.; Liu, Y.; He, C.; Wang, B. Truncated affinity-improved aptamer for selective and sensitive detection of streptomycin in dairy products with label-free electrochemical aptasensor. J. Food Compos. Anal. 2023, 121, 105422. [Google Scholar] [CrossRef]
- Gorgani, L.; Mohammadi, M.; Darzi, G.N.; Raoof, J.B. Electrochemical aptasensor based on bimetallic CuZr-MOF for ultrasensitive detection of miR-21. Sens. Actuators B Chem. 2023, 378, 133194. [Google Scholar] [CrossRef]
- Tığ, G.A.; Pekyardımcı, Ş. An electrochemical sandwich-type aptasensor for determination of lipocalin-2 based on graphene oxide/polymer composite and gold nanoparticles. Talanta 2020, 210, 120666. [Google Scholar]
- Peng, H.; Hui, Y.; Pu, M.; Yang, D.; Zhao, A.; Wang, W.; Wu, S.; Wang, B. Electrochemical aptasensor based on PEI-Fe-MOF/Au@Ag NPs nanocomposites for streptomycin detection in dairy products. J. Food Compos. Anal. 2021, 116, 105091. [Google Scholar] [CrossRef]
- Uhuo, O.V.; Waryo, T.T.; Douman, S.F.; Januarie, K.C.; Nwambaekwe, K.C.; Ndipingwi, M.M.; Ekwere, P.; Iwuoha, E.I. Bioanalytical methods encompassing label-free and labeled tuberculosis aptasensors: A review. Anal. Chim. Acta 2022, 1234, 340326. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.; Li, S.; Li, X.; JiYe, Y.; Mo, Y.; Chen, L.; Zhang, Z.; Wu, H.; Li, M.; Luo, Q. A label-free electrochemical aptasensor based on AuNPs-loaded zeolitic imidazolate framework-8 for sensitive determination of aflatoxin B1. Food Chem. 2022, 384, 132495. [Google Scholar] [CrossRef]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef]
- Li, F.; Wang, X.; Sun, X.; Guo, Y. Multiplex electrochemical aptasensor for detecting multiple antibiotics residues based on carbon fiber and mesoporous carbon-gold nanoparticles. Sens. Actuators B Chem. 2018, 265, 217–226. [Google Scholar] [CrossRef]
- He, H.; Wang, S.-Q.; Han, Z.-Y.; Tian, X.-H.; Zhang, W.-W.; Li, C.-P.; Du, M. Construction of electrochemical aptasensors with Ag(I) metal−organic frameworks toward high-efficient detection of ultra-trace penicillin. Appl. Surf. Sci. 2020, 531, 147342. [Google Scholar] [CrossRef]
- Hui, Y.; Yang, D.; Wei, L.; Pu, M.; Mao, Y.; Chen, X.; Wang, B. Rapid and label-free electrochemical aptasensor based on a palladium nanoparticles/titanium carbide/polyethyleneimine functionalized nitrogen-doped carbon nanotubes composite for amplified detection of streptomycin. Food Chem. 2024, 432, 137271. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, Y.; Xu, W.; Leng, X.; Wang, H.; Guo, Y.; Huang, J. A novel sandwich-type electrochemical aptasensor based on GR-3D Au and aptamer-AuNPs-HRP for sensitive detection of oxytetracycline. Biosens. Bioelectron. 2017, 88, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.Z.; Gu, H.W.; Yi, H.C.; He, Y.Q.; Chen, Y.; Sun, W.Y. Sensitive detection of streptomycin in milk using a hybrid signal enhancement strategy of MOF-based bio-bar code and target recycling. Anal. Chim. Acta 2020, 1125, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hianik, T.; Ostatná, V.; Sonlajtnerova, M.; Grman, I. Influence of ionic strength, pH and aptamer configuration for binding affinity to thrombin. Bioelectrochem 2007, 70, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Kadam, U.S.; Hong, J.C. Advances in aptameric biosensors designed to detect toxic contaminants from food, water, human fluids, and the environment. Trends Environ. Anal. Chem. 2022, 36, e00184. [Google Scholar] [CrossRef]
- Meirinho, S.G.; Dias, L.G.; Peres, A.M.; Rodrigues, L.R. Voltammetric aptasensors for protein disease biomarkers detection: A review. Biotechnol. Adv. 2016, 34, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Grabowska, I.; Zapotoczny, S.; Chlopicki, S. Multiplex electrochemical aptasensors for detection of endothelial dysfunction: Ready for prime time? TrAC Trends Anal. Chem. 2023, 169, 117372. [Google Scholar] [CrossRef]
- Li, F.; Guo, Y.; Wang, X.; Sun, X. Multiplexed aptasensor based on metal ions labels for simultaneous detection of multiple antibiotic residues in milk. Biosens. Bioelectron. 2018, 115, 7–13. [Google Scholar] [CrossRef]
- Wang, Y.; Gong, C.; Zhu, Y.; Wang, Q.; Geng, L. Signal-on electrochemical aptasensor for sensitive detection of sulfamethazine based on carbon quantum dots/tungsten disulfide nanocomposites. Electrochim. Acta 2021, 393, 139054. [Google Scholar] [CrossRef]






| Antibiotic | Technique | Modification | Sample | Detection Range | Limit of Detection (LOD) | Ref. |
|---|---|---|---|---|---|---|
| Tetracycline | DPV | Cu2O@Au | milk | 1.0 nM–1000 µM | 0.16 nM | [50] |
| Streptomycin | EIS | Pencil lead graphite-based electrochemical aptasensor | milk | 10−8–10−16 M | 0.8 × 10−18 M | [51] |
| Oxytetracycline | DPV | 4-carboxyphenyl anchored GCE | milk | 1.0 × 10−9–1.0 × 10−4 g mL−1 | 2.29 × 10−10 g mL−1 | [52] |
| Ampicillin | LSV | MWCNTs | milk | 1.0 × 10−13–1.0 × 10−8 M | 1.0 × 10−13 | [53] |
| Amoxicillin | EIS | TiO2-g-C3N4@Au NPs | wastewater | 0.5–3 nM | 0.2 nM | [54] |
| Ampicillin | DPV | Endonuclease DpnII | milk and water | 0.1–100 nM | 32 pM | [55] |
| Ampicillin | EIS | Co-MOF@TPN-COF | human serum, river water, and milk | 1.0 fg mL−1 –2.0 ng mL−1 | 0.217 fg mL−1 | [56] |
| Tetracycline | SWV | Aptamer cocktail | honey | 0.01–1000 ng mL−1 | 0.0073 ng/mL | [57] |
| Ciprofloxacin | DPV | rGO/PEI/TiO2 | water | 0.003–10.0 μM | 0.7 nM | [58] |
| Streptomycin | DPV | PCNR/GR–Fe3O4–AuNPs | milk | 0.05–200 ng mL−1 | 0.028 ng mL −1 | [59] |
| Kanamycin | DPV | MoS2-Au-HE | milk | 1.0–1.0 × 105 ng L −1 | 0.8 ng L−1 | [60] |
| Sulfaquinoxaline | DPV | AuPd NPs@UiO-66-NH2/CoSe2 | pork | 1.0–100 ng mL−1 | 0.547 pg mL −1 | [61] |
| Tetracycline | EIS | Fe3O4-IL | milk | 1 × 10−9–1 × 10−5 M | 1 × 10−9 M | [62] |
| Tetracycline | SWV | PAN@Cu-BTC | meat | 10 pM–1 µM | 0.32 pM | [63] |
| Tobramycin | DPV | phi29 DNA polymerase and nicking endonuclease Nt.AlwI | milk and water | 10–200 nM | 5.13 nM | [64] |
| Tobramycin | EIS | SnOx@TiO2@mC | human serum and human urine | 0.01–5 ng mL−1 | 6.7 pg mL−1 | [65] |
| Kanamycin | DPV | SA-AuNPs/OMC-CS | milk | 0.1–1000 nM | 0.03569 nM | [66] |
| Streptomycin | EIS | PdNPs/CNT/Chi | milk | 0.10–1500 nM | 18 pM | [67] |
| Chloramphenicol | DPV | Si-Fe/NOMC | eye drop | 1.0–500 μM | 0.03 μM | [68] |
| Ampicillin | EIS | POP | milk | 1.0 × 10–5–5.0 ng mL−1 | 1.33 × 10–6 | [69] |
| Ciprofloxacin | DPV | 3D Au-PAMAM/rGO | raw milk | 1 μM–1.0 nM | 1.0 nM | [70] |
| Kanamycin | DPV | GR-TH/HNP-PtCu | pork meat and chicken | 5 × 10−7–5 × 10−2 μg mL−1 | 0.42 pg mL−1 | [71] |
| Tetracycline | DPV | MBCPE/Fe3O4NPs/OA | drug, milk, honey, and blood serum | 1.0 × 10−10–1.0 × 10−7 M | 2.9 × 10−11 M | [72] |
| Tetracycline | DPV | C-WO3@AuNPs | water, milk, honey, and black tea | 0.1–100 nM | 4.8 × 10−2 nM | [73] |
| Enrofloxacin | SWV | AuPt@h-CeO2/MoS2 | water and milk | 5.0 × 10−6–1.0 × 10−2 ng mL−1 | 1.02 × 10−7 ng mL−1 | [74] |
| Ampicillin | DPV | T7 exonuclease | milk | 0.02–40 nM | 4.0 pM | [75] |
| Chloramphenicol (CAP) and oxytetracycline (OTC) | SWV | NMOF | milk | 10−4–50 nM | CAP 0.033 pM OTC 0.048 pM | [76] |
| Antibiotic | Sequences | Ref. |
|---|---|---|
| Penicillin | 5′-NH2-CTG AATTGGATCTCTCTTCTTGAGCGATCTCCACA-3′ | [91] |
| Streptomycin | 5′-NH2-GGGGTCTGGTGTTCTGCTTTGTTCTGTCGGGTCGT-3′ | [92] |
| Oxytetracycline | 5′-SH-CGACGCACAGTCGCTGGTGCGTACCTGGTTGCCGTTGTG | [93] |
| Kanamycin | 5′-Bio-ACCGCGGGGUUGCGGACCGGGAGCUCCAGC-NH2-3′ | [47] |
| Tobramycin | 5′-Bio-GGCACGAGGUUUAGCUACACUCGUGCC-NH2-3′ | [47] |
| Streptomycin | (SH-cDNA): 5′-ACGACCCGACAGAACAAAGCAGAACACCAGACCCC-SH-3′ Amino-modified STR aptamer (NH2-Apt): 5′-NH2-GGGGTCTGGTGTTCTGCTTTGTTCTGTCGGGTCGT-3′ | [94] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva, D.N.; Pereira, A.C. Relevant Aspects in the Development of Electrochemical Aptasensors for the Determination of Antibiotics—A Review. Electrochem 2023, 4, 553-567. https://doi.org/10.3390/electrochem4040035
da Silva DN, Pereira AC. Relevant Aspects in the Development of Electrochemical Aptasensors for the Determination of Antibiotics—A Review. Electrochem. 2023; 4(4):553-567. https://doi.org/10.3390/electrochem4040035
Chicago/Turabian Styleda Silva, Daniela Nunes, and Arnaldo César Pereira. 2023. "Relevant Aspects in the Development of Electrochemical Aptasensors for the Determination of Antibiotics—A Review" Electrochem 4, no. 4: 553-567. https://doi.org/10.3390/electrochem4040035