Polystyrene Micro- and Nanoplastics (PS MNPs): A Review of Recent Advances in the Use of -Omics in PS MNP Toxicity Studies on Aquatic Organisms
Abstract
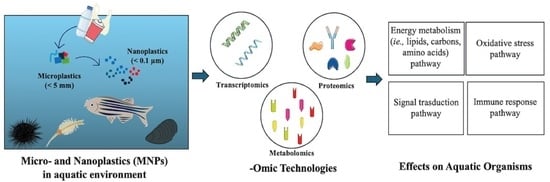
:1. Introduction
2. -Omics Approaches in Studies of PS MNP Toxicity
2.1. Transcriptomics
2.1.1. Effects of PS MNPs on the Transcriptome of Fishes
2.1.2. Effects of PS MNPs on the Transcriptome of Aquatic Invertebrates
2.2. Proteomics
2.2.1. Effects of PS MNPs on the Proteome of Fishes
2.2.2. Effects of PS MNPs on the Proteome of Aquatic Invertebrates
2.3. Metabolomics
2.3.1. Effects of PS MNPs on the Metabolome of Fishes
2.3.2. Effects of PS MNPs on the Metabolome of Aquatic Invertebrates
3. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jambeck, J.R.; Geyer, R.; Wilcox, C.; Siegler, T.R.; Perryman, M.; Andrady, A.; Narayan, R.; Law, K.L. Plastic Waste Inputs from Land into the Ocean. Science 2015, 347, 768–771. [Google Scholar] [CrossRef]
- Gigault, J.; Ter Halle, A.; Baudrimont, M.; Pascal, P.-Y.; Gauffre, F.; Phi, T.-L.; El Hadri, H.; Grassl, B.; Reynaud, S. Current Opinion: What Is a Nanoplastic? Environ. Pollut. 2018, 235, 1030–1034. [Google Scholar] [CrossRef]
- Missawi, O.; Bousserrhine, N.; Zitouni, N.; Maisano, M.; Boughattas, I.; De Marco, G.; Cappello, T.; Belbekhouche, S.; Belbekhouche, S.; Guerrouache, M.; et al. Uptake, Accumulation and Associated Cellular Alterations of Environmental Samples of Microplastics in the Seaworm Hediste diversicolor. J. Hazard. Mater. 2021, 406, 124287. [Google Scholar] [CrossRef]
- Andrady, A.L. Microplastics in the Marine Environment. Mar. Pollut. Bull. 2011, 62, 1596–1605. [Google Scholar] [CrossRef]
- Dawson, A.L.; Kawaguchi, S.; King, C.K.; Townsend, K.A.; King, R.; Huston, W.M.; Bengtson Nash, S.M. Turning Microplastics into Nanoplastics through Digestive Fragmentation by Antarctic Krill. Nat. Commun. 2018, 9, 1001. [Google Scholar] [CrossRef] [PubMed]
- Ekvall, M.T.; Lundqvist, M.; Kelpsiene, E.; Šileikis, E.; Gunnarsson, S.B.; Cedervall, T. Nanoplastics Formed during the Mechanical Breakdown of Daily-Use Polystyrene Products. Nanoscale Adv. 2019, 1, 1055–1061. [Google Scholar] [CrossRef]
- Gigault, J.; El Hadri, H.; Reynaud, S.; Deniau, E.; Grassl, B. Asymmetrical Flow Field Flow Fractionation Methods to Characterize Submicron Particles: Application to Carbon-Based Aggregates and Nanoplastics. Anal. Bioanal. Chem. 2017, 409, 6761–6769. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.; Wagner, M. Characterisation of Nanoplastics during the Degradation of Polystyrene. Chemosphere 2016, 145, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.; Lindeque, P.; Halsband, C.; Galloway, T.S. Microplastics as Contaminants in the Marine Environment: A Review. Mar. Pollut. Bull. 2011, 62, 2588–2597. [Google Scholar] [CrossRef]
- Paul, M.B.; Stock, V.; Cara-Carmona, J.; Lisicki, E.; Shopova, S.; Fessard, V.; Braeuning, A.; Sieg, H.; Böhmert, L. Micro- and Nanoplastics—Current State of Knowledge with the Focus on Oral Uptake and Toxicity. Nanoscale Adv. 2020, 2, 4350–4367. [Google Scholar] [CrossRef]
- Gaylarde, C.C.; Baptista Neto, J.A.; Da Fonseca, E.M. Nanoplastics in Aquatic Systems—Are They More Hazardous than Microplastics? Environ. Pollut. 2021, 272, 115950. [Google Scholar] [CrossRef]
- Klaine, S.J.; Koelmans, A.A.; Horne, N.; Carley, S.; Handy, R.D.; Kapustka, L.; Nowack, B.; Von Der Kammer, F. Paradigms to Assess the Environmental Impact of Manufactured Nanomaterials. Environ. Toxicol. Chem. 2012, 31, 3–14. [Google Scholar] [CrossRef]
- Mattsson, K.; Ekvall, M.T.; Hansson, L.-A.; Linse, S.; Malmendal, A.; Cedervall, T. Altered Behavior, Physiology, and Metabolism in Fish Exposed to Polystyrene Nanoparticles. Environ. Sci. Technol. 2015, 49, 553–561. [Google Scholar] [CrossRef]
- Mattsson, K.; Jocic, S.; Doverbratt, I.; Hansson, L.-A. Nanoplastics in the Aquatic Environment. In Microplastic Contamination in Aquatic Environments; Elsevier: Amsterdam, The Netherlands, 2018; pp. 379–399. ISBN 978-0-12-813747-5. [Google Scholar]
- Shi, C.; Liu, Z.; Yu, B.; Zhang, Y.; Yang, H.; Han, Y.; Wang, B.; Liu, Z.; Zhang, H. Emergence of Nanoplastics in the Aquatic Environment and Possible Impacts on Aquatic Organisms. Sci. Total Environ. 2024, 906, 167404. [Google Scholar] [CrossRef]
- Ferreira, I.; Venâncio, C.; Lopes, I.; Oliveira, M. Nanoplastics and Marine Organisms: What Has Been Studied? Environ. Toxicol. Pharmacol. 2019, 67, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.J. Synthetic Polymers in the Marine Environment: A Rapidly Increasing, Long-Term Threat. Environ. Res. 2008, 108, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Sussarellu, R.; Suquet, M.; Thomas, Y.; Lambert, C.; Fabioux, C.; Pernet, M.E.J.; Le Goïc, N.; Quillien, V.; Mingant, C.; Epelboin, Y.; et al. Oyster Reproduction Is Affected by Exposure to Polystyrene Microplastics. Proc. Natl. Acad. Sci. USA 2016, 113, 2430–2435. [Google Scholar] [CrossRef] [PubMed]
- Della Torre, C.; Bergami, E.; Salvati, A.; Faleri, C.; Cirino, P.; Dawson, K.A.; Corsi, I. Accumulation and Embryotoxicity of Polystyrene Nanoparticles at Early Stage of Development of Sea Urchin Embryos Paracentrotus lividus. Environ. Sci. Technol. 2014, 48, 12302–12311. [Google Scholar] [CrossRef] [PubMed]
- Pinsino, A.; Bergami, E.; Della Torre, C.; Vannuccini, M.L.; Addis, P.; Secci, M.; Dawson, K.A.; Matranga, V.; Corsi, I. Amino-Modified Polystyrene Nanoparticles Affect Signalling Pathways of the Sea Urchin (Paracentrotus lividus) Embryos. Nanotoxicology 2017, 11, 201–209. [Google Scholar] [CrossRef]
- Pitt, J.A.; Kozal, J.S.; Jayasundara, N.; Massarsky, A.; Trevisan, R.; Geitner, N.; Wiesner, M.; Levin, E.D.; Di Giulio, R.T. Uptake, Tissue Distribution, and Toxicity of Polystyrene Nanoparticles in Developing Zebrafish (Danio rerio). Aquat. Toxicol. Amst. Neth. 2018, 194, 185–194. [Google Scholar] [CrossRef]
- Duan, Z.; Duan, X.; Zhao, S.; Wang, X.; Wang, J.; Liu, Y.; Peng, Y.; Gong, Z.; Wang, L. Barrier Function of Zebrafish Embryonic Chorions against Microplastics and Nanoplastics and Its Impact on Embryo Development. J. Hazard. Mater. 2020, 395, 122621. [Google Scholar] [CrossRef]
- Eliso, M.C.; Bergami, E.; Manfra, L.; Spagnuolo, A.; Corsi, I. Toxicity of Nanoplastics during the Embryogenesis of the Ascidian Ciona robusta (Phylum Chordata). Nanotoxicology 2020, 14, 1415–1431. [Google Scholar] [CrossRef] [PubMed]
- Eliso, M.C.; Bergami, E.; Bonciani, L.; Riccio, R.; Belli, G.; Belli, M.; Corsi, I.; Spagnuolo, A. Application of Transcriptome Profiling to Inquire into the Mechanism of Nanoplastics Toxicity during Ciona robusta Embryogenesis. Environ. Pollut. 2023, 318, 120892. [Google Scholar] [CrossRef] [PubMed]
- Balbi, T.; Camisassi, G.; Montagna, M.; Fabbri, R.; Franzellitti, S.; Carbone, C.; Dawson, K.; Canesi, L. Impact of Cationic Polystyrene Nanoparticles (PS-NH2) on Early Embryo Development of Mytilus galloprovincialis: Effects on Shell Formation. Chemosphere 2017, 186, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tallec, K.; Huvet, A.; Di Poi, C.; González-Fernández, C.; Lambert, C.; Petton, B.; Le Goïc, N.; Berchel, M.; Soudant, P.; Paul-Pont, I. Nanoplastics Impaired Oyster Free Living Stages, Gametes and Embryos. Environ. Pollut. 2018, 242, 1226–1235. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Z.; Jiang, Q.; Ye, Y.; Zhao, Y. Effects of Nanoplastic on Cell Apoptosis and Ion Regulation in the Gills of Macrobrachium nipponense. Environ. Pollut. 2022, 300, 118989. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Y.; Zheng, Y.; Feng, Y.; Zhang, W.; Gong, S.; Lin, H.; Gao, P.; Zhang, H. Genome-Wide Identification Glutathione-S-Transferase Gene Superfamily in Daphnia pulex and Its Transcriptional Response to Nanoplastics. Int. J. Biol. Macromol. 2023, 230, 123112. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, C.; Morgana, S.; Ferrando, S.; Bramini, M.; Piazza, V.; Costa, E.; Garaventa, F.; Faimali, M. Effects of Polystyrene Microbeads in Marine Planktonic Crustaceans. Ecotoxicol. Environ. Saf. 2017, 145, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Bergami, E.; Krupinski Emerenciano, A.; González-Aravena, M.; Cárdenas, C.A.; Hernández, P.; Silva, J.R.M.C.; Corsi, I. Polystyrene Nanoparticles Affect the Innate Immune System of the Antarctic Sea Urchin Sterechinus Neumayeri. Polar Biol. 2019, 42, 743–757. [Google Scholar] [CrossRef]
- Qiao, R.; Sheng, C.; Lu, Y.; Zhang, Y.; Ren, H.; Lemos, B. Microplastics Induce Intestinal Inflammation, Oxidative Stress, and Disorders of Metabolome and Microbiome in Zebrafish. Sci. Total Environ. 2019, 662, 246–253. [Google Scholar] [CrossRef]
- Cappello, T.; De Marco, G.; Oliveri Conti, G.; Giannetto, A.; Ferrante, M.; Mauceri, A.; Maisano, M. Time-Dependent Metabolic Disorders Induced by Short-Term Exposure to Polystyrene Microplastics in the Mediterranean Mussel Mytilus galloprovincialis. Ecotoxicol. Environ. Saf. 2021, 209, 111780. [Google Scholar] [CrossRef] [PubMed]
- Murano, C.; Bergami, E.; Liberatori, G.; Palumbo, A.; Corsi, I. Interplay between Nanoplastics and the Immune System of the Mediterranean Sea Urchin Paracentrotus lividus. Front. Mar. Sci. 2021, 8, 647394. [Google Scholar] [CrossRef]
- Murano, C.; Nonnis, S.; Scalvini, F.G.; Maffioli, E.; Corsi, I.; Tedeschi, G.; Palumbo, A. Response to Microplastic Exposure: An Exploration into the Sea Urchin Immune Cell Proteome. Environ. Pollut. 2023, 320, 121062. [Google Scholar] [CrossRef] [PubMed]
- De Marco, G.; Conti, G.O.; Giannetto, A.; Cappello, T.; Galati, M.; Iaria, C.; Pulvirenti, E.; Capparucci, F.; Mauceri, A.; Ferrante, M.; et al. Embryotoxicity of Polystyrene Microplastics in Zebrafish Danio rerio. Environ. Res. 2022, 208, 112552. [Google Scholar] [CrossRef]
- De Marco, G.; Eliso, M.C.; Conti, G.O.; Galati, M.; Billè, B.; Maisano, M.; Ferrante, M.; Cappello, T. Short-Term Exposure to Polystyrene Microplastics Hampers the Cellular Function of Gills in the Mediterranean Mussel Mytilus galloprovincialis. Aquat. Toxicol. 2023, 264, 106736. [Google Scholar] [CrossRef]
- Portugal, J.; Mansilla, S.; Piña, B. Perspectives on the Use of Toxicogenomics to Assess Environmental Risk. Front. Biosci.-Landmark 2022, 27, 294. [Google Scholar] [CrossRef]
- Brockmeier, E.K.; Hodges, G.; Hutchinson, T.H.; Butler, E.; Hecker, M.; Tollefsen, K.E.; Garcia-Reyero, N.; Kille, P.; Becker, D.; Chipman, K.; et al. The Role of Omics in the Application of Adverse Outcome Pathways for Chemical Risk Assessment. Toxicol. Sci. 2017, 158, 252–262. [Google Scholar] [CrossRef]
- Zhang, X.; Xia, P.; Wang, P.; Yang, J.; Baird, D.J. Omics Advances in Ecotoxicology. Environ. Sci. Technol. 2018, 52, 3842–3851. [Google Scholar] [CrossRef]
- Sauer, U.G.; Deferme, L.; Gribaldo, L.; Hackermüller, J.; Tralau, T.; Van Ravenzwaay, B.; Yauk, C.; Poole, A.; Tong, W.; Gant, T.W. The Challenge of the Application of ‘omics Technologies in Chemicals Risk Assessment: Background and Outlook. Regul. Toxicol. Pharmacol. 2017, 91, S14–S26. [Google Scholar] [CrossRef]
- Cui, M.; Cheng, C.; Zhang, L. High-Throughput Proteomics: A Methodological Mini-Review. Lab. Investig. 2022, 102, 1170–1181. [Google Scholar] [CrossRef]
- Wu, Y.; Zeng, J.; Zhang, F.; Zhu, Z.; Qi, T.; Zheng, Z.; Lloyd-Jones, L.R.; Marioni, R.E.; Martin, N.G.; Montgomery, G.W.; et al. Integrative Analysis of Omics Summary Data Reveals Putative Mechanisms Underlying Complex Traits. Nat. Commun. 2018, 9, 918. [Google Scholar] [CrossRef]
- Cappello, T. NMR-Based Metabolomics of Aquatic Organisms. eMagRes 2020, 9, 81–100. [Google Scholar] [CrossRef]
- Ebner, J.N. Trends in the Application of “Omics” to Ecotoxicology and Stress Ecology. Genes 2021, 12, 1481. [Google Scholar] [CrossRef] [PubMed]
- Madeira, C.; Costa, P.M. Proteomics in Systems Toxicology. In Advances in Protein Chemistry and Structural Biology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 127, pp. 55–91. ISBN 978-0-323-85319-4. [Google Scholar]
- Taylor, N.; Gavin, A.; Viant, M. Metabolomics Discovers Early-Response Metabolic Biomarkers That Can Predict Chronic Reproductive Fitness in Individual Daphnia magna. Metabolites 2018, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Aydin, M.E.; Aydin, S.; Tongur, S.; Kara, G.; Kolb, M.; Bahadir, M. Application of Simple and Low-Cost Toxicity Tests for Ecotoxicological Assessment of Industrial Wastewaters. Environ. Technol. 2015, 36, 2825–2834. [Google Scholar] [CrossRef] [PubMed]
- Snape, J.R.; Maund, S.J.; Pickford, D.B.; Hutchinson, T.H. Ecotoxicogenomics: The Challenge of Integrating Genomics into Aquatic and Terrestrial Ecotoxicology. Aquat. Toxicol. 2004, 67, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Suman, A.; Mahapatra, A.; Gupta, P.; Ray, S.S.; Singh, R.K. Polystyrene Microplastics Modulated Bdnf Expression Triggering Neurotoxicity via Apoptotic Pathway in Zebrafish Embryos. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2023, 271, 109699. [Google Scholar] [CrossRef]
- Martin-Folgar, R.; Torres-Ruiz, M.; De Alba, M.; Cañas-Portilla, A.I.; González, M.C.; Morales, M. Molecular Effects of Polystyrene Nanoplastics Toxicity in Zebrafish Embryos (Danio rerio). Chemosphere 2023, 312, 137077. [Google Scholar] [CrossRef]
- Qiang, L.; Cheng, J. Exposure to Microplastics Decreases Swimming Competence in Larval Zebrafish (Danio rerio). Ecotoxicol. Environ. Saf. 2019, 176, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Bao, Z.; Wan, Z.; Fu, Z.; Jin, Y. Polystyrene Microplastic Exposure Disturbs Hepatic Glycolipid Metabolism at the Physiological, Biochemical, and Transcriptomic Levels in Adult Zebrafish. Sci. Total Environ. 2020, 710, 136279. [Google Scholar] [CrossRef]
- Limonta, G.; Mancia, A.; Benkhalqui, A.; Bertolucci, C.; Abelli, L.; Fossi, M.C.; Panti, C. Microplastics Induce Transcriptional Changes, Immune Response and Behavioral Alterations in Adult Zebrafish. Sci. Rep. 2019, 9, 15775. [Google Scholar] [CrossRef]
- Pedersen, A.F.; Meyer, D.N.; Petriv, A.-M.V.; Soto, A.L.; Shields, J.N.; Akemann, C.; Baker, B.B.; Tsou, W.-L.; Zhang, Y.; Baker, T.R. Nanoplastics Impact the Zebrafish (Danio rerio) Transcriptome: Associated Developmental and Neurobehavioral Consequences. Environ. Pollut. Barking Essex 2020, 266, 115090. [Google Scholar] [CrossRef]
- Ašmonaitė, G.; Sundh, H.; Asker, N.; Carney Almroth, B. Rainbow Trout Maintain Intestinal Transport and Barrier Functions Following Exposure to Polystyrene Microplastics. Environ. Sci. Technol. 2018, 52, 14392–14401. [Google Scholar] [CrossRef]
- Lu, C.; Kania, P.W.; Buchmann, K. Particle Effects on Fish Gills: An Immunogenetic Approach for Rainbow Trout and Zebrafish. Aquaculture 2018, 484, 98–104. [Google Scholar] [CrossRef]
- Zhang, Y.T.; Chen, M.; He, S.; Fang, C.; Chen, M.; Li, D.; Wu, D.; Chernick, M.; Hinton, D.E.; Bo, J.; et al. Microplastics Decrease the Toxicity of Triphenyl Phosphate (TPhP) in the Marine Medaka (Oryzias melastigma) Larvae. Sci. Total Environ. 2021, 763, 143040. [Google Scholar] [CrossRef]
- Chen, J.-C.; Chen, M.-Y.; Fang, C.; Zheng, R.-H.; Jiang, Y.-L.; Zhang, Y.-S.; Wang, K.-J.; Bailey, C.; Segner, H.; Bo, J. Microplastics Negatively Impact Embryogenesis and Modulate the Immune Response of the Marine Medaka Oryzias melastigma. Mar. Pollut. Bull. 2020, 158, 111349. [Google Scholar] [CrossRef]
- Varó, I.; Perini, A.; Torreblanca, A.; Garcia, Y.; Bergami, E.; Vannuccini, M.L.; Corsi, I. Time-Dependent Effects of Polystyrene Nanoparticles in Brine Shrimp Artemia franciscana at Physiological, Biochemical and Molecular Levels. Sci. Total Environ. 2019, 675, 570–580. [Google Scholar] [CrossRef]
- Suman, T.Y.; Jia, P.-P.; Li, W.-G.; Junaid, M.; Xin, G.-Y.; Wang, Y.; Pei, D.-S. Acute and Chronic Effects of Polystyrene Microplastics on Brine Shrimp: First Evidence Highlighting the Molecular Mechanism through Transcriptome Analysis. J. Hazard. Mater. 2020, 400, 123220. [Google Scholar] [CrossRef]
- Bergami, E.; Pugnalini, S.; Vannuccini, M.L.; Manfra, L.; Faleri, C.; Savorelli, F.; Dawson, K.A.; Corsi, I. Long-Term Toxicity of Surface-Charged Polystyrene Nanoplastics to Marine Planktonic Species Dunaliella tertiolecta and Artemia franciscana. Aquat. Toxicol. Amst. Neth. 2017, 189, 159–169. [Google Scholar] [CrossRef]
- De Felice, B.; Sugni, M.; Casati, L.; Parolini, M. Molecular, Biochemical and Behavioral Responses of Daphnia magna under Long-Term Exposure to Polystyrene Nanoplastics. Environ. Int. 2022, 164, 107264. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, P.; Cai, M.; Wu, D.; Zhang, M.; Huang, Y.; Zhao, Y. Polystyrene Nanoplastic Exposure Induces Immobilization, Reproduction, and Stress Defense in the Freshwater Cladoceran Daphnia pulex. Chemosphere 2019, 215, 74–81. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; Pérez, E.; Jiang, Q.; Chen, Q.; Jiao, Y.; Huang, Y.; Yang, Y.; Zhao, Y. Polystyrene Nanoplastic Induces Oxidative Stress, Immune Defense, and Glycometabolism Change in Daphnia pulex: Application of Transcriptome Profiling in Risk Assessment of Nanoplastics. J. Hazard. Mater. 2021, 402, 123778. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Z.; Tang, S.; Li, D.; Jiang, Q.; Zhang, T. Transcriptional Response Provides Insights into the Effect of Chronic Polystyrene Nanoplastic Exposure on Daphnia pulex. Chemosphere 2020, 238, 124563. [Google Scholar] [CrossRef]
- Zhu, X.; Teng, J.; Xu, E.G.; Zhao, J.; Shan, E.; Sun, C.; Wang, Q. Toxicokinetics and Toxicodynamics of Plastic and Metallic Nanoparticles: A Comparative Study in Shrimp. Environ. Pollut. 2022, 312, 120069. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, H.; Luan, L.; Luo, X.; Wang, X.; Lu, H.; Li, Y.; Wen, L.; Li, F.; Zhao, J. Functionalized Polystyrene Nanoplastic-Induced Energy Homeostasis Imbalance and the Immunomodulation Dysfunction of Marine Clams (Meretrix Meretrix) at Environmentally Relevant Concentrations. Environ. Sci. Nano 2021, 8, 2030–2048. [Google Scholar] [CrossRef]
- Capolupo, M.; Franzellitti, S.; Valbonesi, P.; Lanzas, C.S.; Fabbri, E. Uptake and Transcriptional Effects of Polystyrene Microplastics in Larval Stages of the Mediterranean Mussel Mytilus galloprovincialis. Environ. Pollut. 2018, 241, 1038–1047. [Google Scholar] [CrossRef]
- Auguste, M.; Lasa, A.; Balbi, T.; Pallavicini, A.; Vezzulli, L.; Canesi, L. Impact of Nanoplastics on Hemolymph Immune Parameters and Microbiota Composition in Mytilus galloprovincialis. Mar. Environ. Res. 2020, 159, 105017. [Google Scholar] [CrossRef]
- Gardon, T.; Morvan, L.; Huvet, A.; Quillien, V.; Soyez, C.; Le Moullac, G.; Le Luyer, J. Microplastics Induce Dose-Specific Transcriptomic Disruptions in Energy Metabolism and Immunity of the Pearl Oyster Pinctada Margaritifera. Environ. Pollut. 2020, 266, 115180. [Google Scholar] [CrossRef]
- Capanni, F.; Greco, S.; Tomasi, N.; Giulianini, P.G.; Manfrin, C. Orally Administered Nano-Polystyrene Caused Vitellogenin Alteration and Oxidative Stress in the Red Swamp Crayfish (Procambarus clarkii). Sci. Total Environ. 2021, 791, 147984. [Google Scholar] [CrossRef]
- Magyary, I. Recent Advances and Future Trends in Zebrafish Bioassays for Aquatic Ecotoxicology. Ecocycles 2018, 4, 12–18. [Google Scholar] [CrossRef]
- Benson, N.U.; Agboola, O.D.; Fred-Ahmadu, O.H.; De-la-Torre, G.E.; Oluwalana, A.; Williams, A.B. Micro(Nano)Plastics Prevalence, Food Web Interactions, and Toxicity Assessment in Aquatic Organisms: A Review. Front. Mar. Sci. 2022, 9, 851281. [Google Scholar] [CrossRef]
- Rosner, A.; Ballarin, L.; Barnay-Verdier, S.; Borisenko, I.; Drago, L.; Drobne, D.; Concetta Eliso, M.; Harbuzov, Z.; Grimaldi, A.; Guy-Haim, T.; et al. A Broad-taxa Approach as an Important Concept in Ecotoxicological Studies and Pollution Monitoring. Biol. Rev. 2023, 99, 131–176. [Google Scholar] [CrossRef]
- Yin, J.; Long, Y.; Xiao, W.; Liu, D.; Tian, Q.; Li, Y.; Liu, C.; Chen, L.; Pan, Y. Ecotoxicology of Microplastics in Daphnia: A Review Focusing on Microplastic Properties and Multiscale Attributes of Daphnia. Ecotoxicol. Environ. Saf. 2023, 249, 114433. [Google Scholar] [CrossRef]
- Corsi, I.; Bellingeri, A.; Eliso, M.C.; Grassi, G.; Liberatori, G.; Murano, C.; Sturba, L.; Vannuccini, M.L.; Bergami, E. Eco-Interactions of Engineered Nanomaterials in the Marine Environment: Towards an Eco-Design Framework. Nanomaterials 2021, 11, 1903. [Google Scholar] [CrossRef]
- Lee, K.W.; Park, H.G.; Lee, S.-M.; Kang, H.-K. Effects of Diets on the Growth of the Brackish Water Cyclopoid Copepod Paracyclopina nana Smirnov. Aquaculture 2006, 256, 346–353. [Google Scholar] [CrossRef]
- Pinto, C.S.C.; Souza-Santos, L.P.; Santos, P.J.P. Development and Population Dynamics of Tisbe biminiensis (Copepoda: Harpacticoida) Reared on Different Diets. Aquaculture 2001, 198, 253–267. [Google Scholar] [CrossRef]
- Sun, B.; Fleeger, J.W. Sustained Mass Culture of Amphiascoides atopus a Marine Harpacticoid Copepod in a Recirculating System. Aquaculture 1995, 136, 313–321. [Google Scholar] [CrossRef]
- Botterell, Z.L.R.; Beaumont, N.; Dorrington, T.; Steinke, M.; Thompson, R.C.; Lindeque, P.K. Bioavailability and Effects of Microplastics on Marine Zooplankton: A Review. Environ. Pollut. 2019, 245, 98–110. [Google Scholar] [CrossRef]
- Dupree, E.J.; Jayathirtha, M.; Yorkey, H.; Mihasan, M.; Petre, B.A.; Darie, C.C. A Critical Review of Bottom-Up Proteomics: The Good, the Bad, and the Future of This Field. Proteomes 2020, 8, 14. [Google Scholar] [CrossRef]
- Duong, V.-A.; Park, J.-M.; Lee, H. Review of Three-Dimensional Liquid Chromatography Platforms for Bottom-Up Proteomics. Int. J. Mol. Sci. 2020, 21, 1524. [Google Scholar] [CrossRef]
- Chen, G.; Pramanik, B.N. LC-MS for Protein Characterization: Current Capabilities and Future Trends. Expert Rev. Proteom. 2008, 5, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Liu, Y.; Pramanik, B.N. LC/MS Analysis of Proteins and Peptides in Drug Discovery. In HPLC for Pharmaceutical Scientists; Kazakevich, Y., LoBrutto, R., Eds.; Wiley: Hoboken, NJ, USA, 2007; pp. 837–899. ISBN 978-0-471-68162-5. [Google Scholar]
- Chen, G.; Pramanik, B.N.; Liu, Y.; Mirza, U.A. Applications of LC/MS in Structure Identifications of Small Molecules and Proteins in Drug Discovery. J. Mass Spectrom. 2007, 42, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Pramanik, B.N. Application of LC/MS to Proteomics Studies: Current Status and Future Prospects. Drug Discov. Today 2009, 14, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Gajahin Gamage, N.T.; Miyashita, R.; Takahashi, K.; Asakawa, S.; Senevirathna, J.D.M. Proteomic Applications in Aquatic Environment Studies. Proteomes 2022, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Tian, L.; Gao, G.; Peng, S.; Zhang, J.; Wu, D.; Huang, J.; Hua, Q.; Lu, T.; Zhong, L.; et al. Inhibitory Effects of Polystyrene Microplastics on Caudal Fin Regeneration in Zebrafish Larvae. Environ. Pollut. 2020, 266, 114664. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lai, H.; Huang, J.; Sun, L.; Mennigen, J.A.; Wang, Q.; Liu, Y.; Jin, Y.; Tu, W. Polystyrene Microplastics Decrease F–53B Bioaccumulation but Induce Inflammatory Stress in Larval Zebrafish. Chemosphere 2020, 255, 127040. [Google Scholar] [CrossRef]
- Jeong, C.-B.; Won, E.-J.; Kang, H.-M.; Lee, M.-C.; Hwang, D.-S.; Hwang, U.-K.; Zhou, B.; Souissi, S.; Lee, S.-J.; Lee, J.-S. Microplastic Size-Dependent Toxicity, Oxidative Stress Induction, and p-JNK and p-P38 Activation in the Monogonont Rotifer (Brachionus koreanus). Environ. Sci. Technol. 2016, 50, 8849–8857. [Google Scholar] [CrossRef]
- Trotter, B.; Wilde, M.V.; Brehm, J.; Dafni, E.; Aliu, A.; Arnold, G.J.; Fröhlich, T.; Laforsch, C. Long-Term Exposure of Daphnia magna to Polystyrene Microplastic (PS-MP) Leads to Alterations of the Proteome, Morphology and Life-History. Sci. Total Environ. 2021, 795, 148822. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, T.; Liu, X.; Gu, X.; Li, D.; Yin, J.; Jiang, Q.; Zhang, W. Changes in Life-History Traits, Antioxidant Defense, Energy Metabolism and Molecular Outcomes in the Cladoceran Daphnia pulex after Exposure to Polystyrene Microplastics. Chemosphere 2022, 308, 136066. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; Sepúlveda, M.S.; Jiang, Q.; Jiao, Y.; Chen, Q.; Huang, Y.; Tian, J.; Zhao, Y. Development of an Adverse Outcome Pathway for Nanoplastic Toxicity in Daphnia pulex Using Proteomics. Sci. Total Environ. 2021, 766, 144249. [Google Scholar] [CrossRef] [PubMed]
- Magni, S.; Della Torre, C.; Garrone, G.; D’Amato, A.; Parenti, C.C.; Binelli, A. First Evidence of Protein Modulation by Polystyrene Microplastics in a Freshwater Biological Model. Environ. Pollut. 2019, 250, 407–415. [Google Scholar] [CrossRef]
- Duan, Y.; Xiong, D.; Wang, Y.; Zhang, Z.; Li, H.; Dong, H.; Zhang, J. Toxicological Effects of Microplastics in Litopenaeus vannamei as Indicated by an Integrated Microbiome, Proteomic and Metabolomic Approach. Sci. Total Environ. 2021, 761, 143311. [Google Scholar] [CrossRef]
- Jeong, C.-B.; Kang, H.-M.; Lee, M.-C.; Kim, D.-H.; Han, J.; Hwang, D.-S.; Souissi, S.; Lee, S.-J.; Shin, K.-H.; Park, H.G.; et al. Adverse Effects of Microplastics and Oxidative Stress-Induced MAPK/Nrf2 Pathway-Mediated Defense Mechanisms in the Marine Copepod Paracyclopina nana. Sci. Rep. 2017, 7, 41323. [Google Scholar] [CrossRef]
- Zhang, C.; Jeong, C.-B.; Lee, J.-S.; Wang, D.; Wang, M. Transgenerational Proteome Plasticity in Resilience of a Marine Copepod in Response to Environmentally Relevant Concentrations of Microplastics. Environ. Sci. Technol. 2019, 53, 8426–8436. [Google Scholar] [CrossRef]
- Oliver, S. Systematic Functional Analysis of the Yeast Genome. Trends Biotechnol. 1998, 16, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Viant, M.R.; Tjeerdema, R.S. Metabolomics: Methodologies and Applications in the Environmental Sciences. J. Pestic. Sci. 2006, 31, 245–251. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond Biomarkers and towards Mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Nagana Gowda, G.A.; Raftery, D. NMR-Based Metabolomics. In Cancer Metabolomics; Hu, S., Ed.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2021; Volume 1280, pp. 19–37. ISBN 978-3-030-51651-2. [Google Scholar]
- Gowda, G.A.N.; Djukovic, D. Overview of Mass Spectrometry-Based Metabolomics: Opportunities and Challenges. In Mass Spectrometry in Metabolomics; Raftery, D., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2014; Volume 1198, pp. 3–12. ISBN 978-1-4939-1257-5. [Google Scholar]
- Idle, J.R.; Gonzalez, F.J. Metabolomics. Cell Metab. 2007, 6, 348–351. [Google Scholar] [CrossRef] [PubMed]
- De Marco, G.; Billè, B.; Brandão, F.; Galati, M.; Pereira, P.; Cappello, T.; Pacheco, M. Differential Cell Metabolic Pathways in Gills and Liver of Fish (White Seabream Diplodus sargus) Coping with Dietary Methylmercury Exposure. Toxics 2023, 11, 181. [Google Scholar] [CrossRef]
- Brandão, F.; Cappello, T.; Raimundo, J.; Santos, M.A.; Maisano, M.; Mauceri, A.; Pacheco, M.; Pereira, P. Unravelling the Mechanisms of Mercury Hepatotoxicity in Wild Fish (Liza aurata) through a Triad Approach: Bioaccumulation, Metabolomic Profiles and Oxidative Stress. Metallomics 2015, 7, 1352–1363. [Google Scholar] [CrossRef]
- Cappello, T.; Maisano, M.; Giannetto, A.; Parrino, V.; Mauceri, A.; Fasulo, S. Neurotoxicological Effects on Marine Mussel Mytilus galloprovincialis Caged at Petrochemical Contaminated Areas (Eastern Sicily, Italy): 1H NMR and Immunohistochemical Assays. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2015, 169, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Cappello, T.; Brandão, F.; Guilherme, S.; Santos, M.A.; Maisano, M.; Mauceri, A.; Canário, J.; Pacheco, M.; Pereira, P. Insights into the Mechanisms Underlying Mercury-Induced Oxidative Stress in Gills of Wild Fish (Liza aurata) Combining 1 H NMR Metabolomics and Conventional Biochemical Assays. Sci. Total Environ. 2016, 548–549, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Cappello, T.; Maisano, M.; Mauceri, A.; Fasulo, S. 1 H NMR-Based Metabolomics Investigation on the Effects of Petrochemical Contamination in Posterior Adductor Muscles of Caged Mussel Mytilus galloprovincialis. Ecotoxicol. Environ. Saf. 2017, 142, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Cappello, T.; Giannetto, A.; Parrino, V.; De Marco, G.; Mauceri, A.; Maisano, M. Food Safety Using NMR-Based Metabolomics: Assessment of the Atlantic Bluefin Tuna, Thunnus thynnus, from the Mediterranean Sea. Food Chem. Toxicol. 2018, 115, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Missawi, O.; Venditti, M.; Cappello, T.; Zitouni, N.; Marco, G.D.; Boughattas, I.; Bousserrhine, N.; Belbekhouche, S.; Minucci, S.; Maisano, M.; et al. Autophagic Event and Metabolomic Disorders Unveil Cellular Toxicity of Environmental Microplastics on Marine Polychaete Hediste Diversicolor. Environ. Pollut. 2022, 302, 119106. [Google Scholar] [CrossRef] [PubMed]
- Vignet, C.; Cappello, T.; Fu, Q.; Lajoie, K.; De Marco, G.; Clérandeau, C.; Mottaz, H.; Maisano, M.; Hollender, J.; Schirmer, K.; et al. Imidacloprid Induces Adverse Effects on Fish Early Life Stages That Are More Severe in Japanese Medaka (Oryzias latipes) than in Zebrafish (Danio rerio). Chemosphere 2019, 225, 470–478. [Google Scholar] [CrossRef]
- Zitouni, N.; Cappello, T.; Missawi, O.; Boughattas, I.; De Marco, G.; Belbekhouche, S.; Mokni, M.; Alphonse, V.; Guerbej, H.; Bousserrhine, N.; et al. Metabolomic Disorders Unveil Hepatotoxicity of Environmental Microplastics in Wild Fish Serranus Scriba (Linnaeus 1758). Sci. Total Environ. 2022, 838, 155872. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.-D.; Wang, J.-S.; Li, M.-H.; Liu, Y.; Chen, T.; Jia, A.-Q. 1H NMR Based Metabolomics Approach to Study the Toxic Effects of Herbicide Butachlor on Goldfish (Carassius auratus). Aquat. Toxicol. 2015, 159, 69–80. [Google Scholar] [CrossRef]
- Wan, Z.; Wang, C.; Zhou, J.; Shen, M.; Wang, X.; Fu, Z.; Jin, Y. Effects of Polystyrene Microplastics on the Composition of the Microbiome and Metabolism in Larval Zebrafish. Chemosphere 2019, 217, 646–658. [Google Scholar] [CrossRef]
- Kaloyianni, M.; Bobori, D.C.; Xanthopoulou, D.; Malioufa, G.; Sampsonidis, I.; Kalogiannis, S.; Feidantsis, K.; Kastrinaki, G.; Dimitriadi, A.; Koumoundouros, G.; et al. Toxicity and Functional Tissue Responses of Two Freshwater Fish after Exposure to Polystyrene Microplastics. Toxics 2021, 9, 289. [Google Scholar] [CrossRef]
- Wang, C.; Hou, M.; Shang, K.; Wang, H.; Wang, J. Microplastics (Polystyrene) Exposure Induces Metabolic Changes in the Liver of Rare Minnow (Gobiocypris rarus). Molecules 2022, 27, 584. [Google Scholar] [CrossRef]
- Pang, M.; Wang, Y.; Tang, Y.; Dai, J.; Tong, J.; Jin, G. Transcriptome Sequencing and Metabolite Analysis Reveal the Toxic Effects of Nanoplastics on Tilapia after Exposure to Polystyrene. Environ. Pollut. 2021, 277, 116860. [Google Scholar] [CrossRef]
- Ding, J.; Huang, Y.; Liu, S.; Zhang, S.; Zou, H.; Wang, Z.; Zhu, W.; Geng, J. Toxicological Effects of Nano- and Micro-Polystyrene Plastics on Red Tilapia: Are Larger Plastic Particles More Harmless? J. Hazard. Mater. 2020, 396, 122693. [Google Scholar] [CrossRef]
- Usman, S.; Razis, A.F.A.; Shaari, K.; Azmai, M.N.A.; Saad, M.Z.; Isa, N.M.; Nazarudin, M.F. Polystyrene Microplastics Induce Gut Microbiome and Metabolome Changes in Javanese Medaka Fish (Oryzias javanicus Bleeker, 1854). Toxicol. Rep. 2022, 9, 1369–1379. [Google Scholar] [CrossRef]
- Ye, G.; Zhang, X.; Liu, X.; Liao, X.; Zhang, H.; Yan, C.; Lin, Y.; Huang, Q. Polystyrene Microplastics Induce Metabolic Disturbances in Marine Medaka (Oryzias melastigmas) Liver. Sci. Total Environ. 2021, 782, 146885. [Google Scholar] [CrossRef]
- Sun, X.; Wang, X.; Booth, A.M.; Zhu, L.; Sui, Q.; Chen, B.; Qu, K.; Xia, B. New Insights into the Impact of Polystyrene Micro/Nanoplastics on the Nutritional Quality of Marine Jacopever (Sebastes schlegelii). Sci. Total Environ. 2023, 903, 166560. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-K.; Yang, B.-K.; Zhang, C.-N.; Xu, S.-X.; Sun, P. Effects of Polystyrene Microplastics Acute Exposure in the Liver of Swordtail Fish (Xiphophorus helleri) Revealed by LC-MS Metabolomics. Sci. Total Environ. 2022, 850, 157772. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Zhao, J.; Teng, J.; Ren, J.; Shan, E.; Zhu, X.; Zhang, W.; Wang, L.; Hou, C.; Wang, Q. Combined Effects of Salinity and Polystyrene Microplastics Exposure on the Pacific Oysters Crassostrea Gigas: Oxidative Stress and Energy Metabolism. Mar. Pollut. Bull. 2023, 193, 115153. [Google Scholar] [CrossRef] [PubMed]
- Kelpsiene, E.; Cedervall, T.; Malmendal, A. Metabolomics-Based Analysis in Daphnia magna after Exposure to Low Environmental Concentrations of Polystyrene Nanoparticles. Environ. Sci. Nano 2023, 10, 1858–1866. [Google Scholar] [CrossRef]
- Zeng, Y.; Deng, B.; Kang, Z.; Araujo, P.; Mjøs, S.A.; Liu, R.; Lin, J.; Yang, T.; Qu, Y. Tissue Accumulation of Polystyrene Microplastics Causes Oxidative Stress, Hepatopancreatic Injury and Metabolome Alterations in Litopenaeus vannamei. Ecotoxicol. Environ. Saf. 2023, 256, 114871. [Google Scholar] [CrossRef] [PubMed]


| FISHES | ||||||||
|---|---|---|---|---|---|---|---|---|
| Method Used | Organism Tested | PS MNPs Size | Concentration Tested | Time of Exposure | Organ/Tissue Target | Life Stage | Effect | References |
| qRT-PCR | Danio rerio (zebrafish) | 10 µm | 200 particles/mL | 120 hpf | Whole organism | Larvae | ↑ sod1, sod2, cat, gst and cyp | [35] |
| qRT-PCR | Danio rerio (zebrafish) | 500 nm | 0.1, 1 and 10 ppm | 96 hpf | Whole organism | Larvae | ↑ p53, cas-3 and cas-9; ↓ bcl and bdnf | [49] |
| qRT-PCR | Danio rerio (zebrafish) | 30 nm | 0.1, 0.5 and 3 ppm | 120 hpf | Whole organism | Larvae | ↑ sod1, sod2, cas-1, cas-8, and il1β; ↓ hsp70, bcl-2, ache, DNA repair genes gadd45α and rad51 | [50] |
| qRT-PCR | Danio rerio (zebrafish) | 1 µm | 1000 µg/L (around 1.91 × 107 particles/L) | 96 hpf | Whole organism | Larvae | ↑ il1β; ↓ cat | [51] |
| qRT-PCR and RNA-seq | Danio rerio (zebrafish) | 5 μm | 20–100 μg/L | 21 d | Liver | Adult | ↑ aco and fabp6; ↓ cpt1, ppar-α, acc1, fas, srebp1α, ppar-α. KEGG pathways analysis revealed carbon, lipid and amino acid metabolism effect | [52] |
| RNA-seq | Danio rerio (zebrafish) | size ranging from 25 to 90 µm | 100–1000 µg/L | 20 d | Liver | Adult | Alteration in pathways related to immune response and lipid metabolism, i.e., sterol biosynthetic process, steroid metabolic process and fatty acid metabolic process | [53] |
| RNA-seq | Danio rerio (zebrafish) | 50 and 200 nm, | 100–1000 ppb | 5 d | Whole organism | Larvae | Nervous system development and function pathways | [54] |
| qRT-PCR | Oncorhynchus mykiss (rainbow trout) | 100–400 µm | 500–2411 particles/fish/day | 4 w | Intestine | Adult | No change in immune response related genes | [55] |
| qRT-PCR | Oncorhynchus mykiss (rainbow trout) | 0.2, 1, 20, 40 and 90 μm | 2 × 105 particles/L | 2 h | Gills | Adult | ↑ ifnγ gene exposed to 0.2 and 40 μm beads; ↓ il1β (bead size 1 μm), s100a (bead size 40 μm) and saa (1, 40 and 90 μm) | [56] |
| qRT-PCR | Oryzias melastigma (marine medaka) | plain PS, carboxylated PS: PS-COOH and aminated PS: PS-NH2 with a size of 1 μm | 0.02 mg/L | 7 d | Whole organism | Larvae | ↓ cox1 and cox2 by PS, PS-COOH and PS-NH2; ↓ cyp1a, multifunction gene (ATPase) by PS-NH2 and PS-COOH, respectively. No impairment of oxidative stress genes in all the treatments | [57] |
| RNA-seq and qRT-PCR | Oryzias melastigma (marine medaka) | 0.05, 0.50, and 6.00 μm | 0.1; 1 × 103; 1 × 106 particles/mL | 19 d | Whole organism | Larvae | ↓ inflammatory and immune-related signaling pathways (Hippo, B cell receptor, RIG-I-like receptor, and inflammatory mediator regulation of the TRP-channels-signaling pathway); heart development (↓ gata4 and nkx2.5, and ↑ bmp4) hatching enzyme (hce and lce) | [58] |
| AQUATIC INVERTEBRATES | ||||||||
| qRT-PCR | Artemia franciscana (brine shrimp) | 50 nm PS-NH2 | 1 μg/mL | 14 d | Whole organism | Adult | ↑ clap and cstb genes | [59] |
| RNA-seq | Artemia franciscana (brine shrimp) | 5 µm | 1 mg/L | 14 d | Whole organism | Adult | KEGG enrichment analysis mapped into arrhythmogenic right ventricular cardiomyopathy, viral myocarditis, hypertrophic cardiomyopathy, phagosome, fluid shear stress, atherosclerosis and regulations of actin cytoskeleton, with most of the DEGs correlated with ROS activity and apoptosis activity | [60] |
| qRT-PCR | Artemia franciscana (brine shrimp) | 50 nm PS-NH2 | 1 μg/mL | 48 h and 14 d | Whole organism | Neonates and adult | Time-dependent ↑ clap and cstb genes and hsp60 and hsp70 | [61] |
| RNA-seq | Ciona robusta (ascidian) | 50 nm PS-NH2 | 10 and 15 μg/mL | 22 hpf | Whole organism | Embryos | ↓ genes involved in glutathione metabolism (glutamate--cysteine ligase catalytic subunit-like transcript variant X1 and X2; glutathione S-transferase omega-1-like), immune defense (integumentary mucin C.1-like transcript variant; mucin-5AC transcript variant; interferon-induced protein 44-like; plasminogen-like), nervous system (acetylcholinesterase-like; sco-spondin), transport by aquaporins (aquaporin-like) and energy metabolism (succinate--CoA ligase [ADP/GDP-forming] subunit alpha mitochondrial-like; 6-phosphofructo-2-kinase/fructose-26-bisphosphatase 1 transcript variant; glycoside hydrolase transcript variant) | [24] |
| qRT-PCR | Daphnia magna (water flea) | 50 nm | 0.05, 0.5 μg/L | 21 d | Whole organism | Adult | ↓ cat after exposure of 21 d to 0.5 μg/mL | [62] |
| qRT-PCR | Daphnia pulex (water flea) | 75 nm | 0.1, 0.5, 1, 2 mg/L | 21 d | Whole organism | Adult | Sod, gst, gpx and cat initially ↑ and then inhibited. ↑ hsp in all the treatment groups | [63] |
| RNA-seq | Daphnia pulex (water flea) | ~70 nm | 1 mg/L (5.32 × 108 particles/mL) | 96 h | Whole organism | Neonates | Alterations in oxidative stress (arachidonic acid metabolism, glutathione metabolism, and porphyrin and chlorophyll metabolism), immune response (drug metabolism–cyp450 and other enzymes, metabolism of xenobiotics by cyp450, glutathione metabolism, hippo signaling pathway, and adherens junction) and energy metabolism pathways (starch and sucrose metabolism, pentose and glucuronate interconversions, galactose metabolism, fructose and mannose metabolism, carbohydrate digestion and absorption, and glycolysis/gluconeogenesis) | [64] |
| RNA-seq | Daphnia pulex (water flea) | 75 nm | 1 mg/L | 21 d | Whole organism | Adult | Alteration in genes involved in chitin metabolism, trehalose transport and metabolism, growth-related genes, long-chain fatty acids metabolism, defense mechanisms, and sex differentiation | [65] |
| qRT-PCR | Litopenaeus vannamei (whiteleg shrimp) | 100 nm | 200 and 2000 mg/kg | 14 and 28 d | Hepatopancreas | Adult | ↑ Beta-glucan binding protein, LPS/β-glucan binding protein, and hsp90 genes. ↑ TLR gene | [66] |
| RNA-seq | Meretrix meretrix (marine clam) | 100 nm PS-NH2 200 nm PS-COOH | 2 mg/L | 7 d | Digestive gland | Adult | ↑ energy homeostasis imbalance (e.g., lipid metabolism, PPAR signaling pathway, protein digestion and absorption, pyruvate metabolism, and glycolysis; Impairment of immune system (e.g., NLRs, NF-κB signaling pathway, TLR signaling pathway, phagosome, lysosome, and apoptosis) | [67] |
| qRT-PCR | Mytilus galloprovincialis (Mediterranean mussel) | 50 nm PS-NH2 | 0.150 mg/L | 24 and 48 hpf | Whole organism | Embryos | ↓ cs, ca, and ep genes | [25] |
| qRT-PCR | Mytilus galloprovincialis (Mediterranean mussel) | 3 µm | 50–500 particles/mL | 24 and 48 hpf | Whole organism | Embryos | ↑ ep, ca, and cs genes; ↑ mytc and mytb genes; ↓ gusb, hex, ctsl genes | [68] |
| qRT-PCR | Mytilus galloprovincialis (Mediterranean mussel) | 50 nm PS-NH2 | 10 μg/L | First exposure 24 h, rest period 72 h, second exposure 24 h | Hemocytes | Adult | ↑ epp, lyso, amps, mytb, mytc, frep | [69] |
| qRT-PCR | Paracentrotus lividus (sea urchin) | 50 nm PS-NH2 | 3 μg/mL | 24 and 48 hpf | Whole organism | Embryos | ↑ cas8 | [19] |
| qRT-PCR | Paracentrotus lividus (sea urchin) | 50 nm PS-NH2 | 3 and 4 μg/mL | 24 and 48 hpf | Whole organism | Embryos | ↑ hsp70, p38 Mapk, univin and cas8 | [20] |
| RNA-seq | Pinctada margaritifera (black-lip pearl oyster) | 6 and 10 µm | 0.25–2.5–25 μg/L | 2 months | Mantle | Adult | Alteration in energy, stress, and immune-related genes. ↓ cyp2d11, gst1, sult1c4 and abcb1, cel and actin gene | [70] |
| RNA-seq | Procambarus clarkia (red swamp crayfish) | 0.10 μm | 1.4 × 1011 particles/L | 72 h | Hemocytes and hepatopancreas | Adult | In hemocytes, ↑ 8 DEGs involved in gene transcription and translation. In hepatopancreas, differential expression of only 3 genes (cyp49a1 and two unknown genes) | [71] |
| qRT-PCR | Sterechinus neumayeri (Antarctic sea urchin) | 40 nm PS-COOH 50 nm PS-NH2 | 1 and 5 μg/mL | 6 and 24 h | Coelomocytes | Adult | ↑ antioxidant genes at 1 µg/mL PS-COOH. ↓ sod at 1 µg/mL PS-NH2, ↑ mt at both the concentrations tested PS-NH2. ↑ the immune-related gene NF-kB and LBP/BPI by PS-NH2 | [30] |
| FISHES | ||||||||
|---|---|---|---|---|---|---|---|---|
| Method Used | Organism Tested | PS MNPs Size | Concentration Tested | Time of Exposure | Organ/Tissue Target | Life Stage | Effect | References |
| ELISA | Danio rerio (zebrafish) | 50 and 500 nm | 0.1, 1, 10 mg/L | 12 and 72 hpa | Whole organism | Larvae | Slight ↑ Il-10, Tnf-α and Nf-κB at the lowest dose. ↓ Il-10, Tnf-α, FgF20a and Nf-κB at the higher dose | [88] |
| WB | Danio rerio (zebrafish) | 5 µm | 50 ng/mL | 7 d | Whole organism | Larvae | ↑ iNOS and Nf-κB | [89] |
| AQUATIC INVERTEBRATES | ||||||||
| WB | Brachionus koreanus (marine rotifer) | 0.05 and 0.5 μm | 10 µg/mL | 24 h | Whole organism | Neonates | ↑ phosphorylation of JNK and p38 Mapk related to ↑ ROS level | [90] |
| LC-MS/MS | Crassostrea gigas (Pacific oyster) | 2 and 6 μm | 0.023 mg/L | 60 d | Oocytes | Gamets | ↓ arginine kinase and ↑ severin | [18] |
| LC-MS/MS | Daphnia magna (water flea) | Mean particle size 13.03 ± 7.75 μm | 101.6 mg/L | 19 d | Whole organism | Adult | Changes in 41 proteins, mostly those related to sulfation, chitin-binding and cuticle’s structural integrity. The less abundant proteins are related to pigment binding, response to stimuli, response to ROS, response to oxidative stress, and response to oxygen-containing compound | [91] |
| RPLC/MS | Daphnia pulex (water flea) | 500 nm | 1 mg/L | 14 d | Whole organism | Adult | Changes in 89 proteins, including those involved in P-body assembly, nuclear-transcribed mRNA catabolic process, ATP-dependent chromatin remodeling, energy metabolism and unfolded protein responses | [92] |
| LC-MS/MS | Daphnia pulex (water flea) | Mean particle size 71.18 nm | 0.5–2 mg/L | 21 d | Whole organism | Adult | 327 proteins ↓, including those involved in cell signaling, immune function, detoxification, energy metabolism, ECM-receptor interaction pathways, and glutathione metabolism | [93] |
| nLC-MS/MS | Dreissena polymorpha (Zebra mussel) | 1 and 10 μm size | 4 × 106 particles/L mixtures | 6 d | Gills | Adult | 78 proteins ↓ and 18 proteins not expressed. Effect of catalytic activity (27%), nucleotide binding, proteins involved in structural molecule activity (12%) and protein binding (11%), proteins related to RNA (5%) and metal ion (4%) bindings | [94] |
| LC-MS/MS | Litopenaeus vannamei (Pacific white shirmp) | 100–200 μm | 1 mg/L | 14 d | Haemolymph | Adult | 47 proteins ↓, including those belonging to extracellular, plasma membrane and lysosomal localization, and related to T cell receptor signaling pathway, epithelial cell signaling in Helicobacter pylori infection, and phospholipase D signaling pathway | [95] |
| Nano HPLC MS/MS | Paracentrotus lividus (sea urchin) | 45 μm | 10, 50, 103, 104 particles/L | 72 h | Coelomocytes | Adult | ↑ proteins involved in endosome transport via multivesicular body sorting pathway and in establishment of protein localisation, and proteins involved in catabolic processes | [34] |
| WB | Paracyclopina nana (marine copepod) | 0.05 μm | 10 µg/mL | 24 h | Whole organism | Neonates | Oxidative stress induction (↑ ROS level) and ↑ phosphorilation of the proteins p38 Mapk, ERK and Nrf2 | [96] |
| LC-MS/MS | Tigriopus japonicus (marine copepod) | 6 μm | 0.23 mg/L | Two generations (F1 and F2) | Whole organism | Adult | ↑ proteins involved in several cellular biosynthesis and ↓ cellular energy storage in F1 generation. Transgenerational proteome plasticity in F2 generation with elevated energy metabolism and stress related defense | [97] |
| FISHES | ||||||||
|---|---|---|---|---|---|---|---|---|
| Method Used | Organism Tested | PS MNPs Size | Concentration Tested | Time of Exposure | Organ/Tissue Target | Life Stage | Effect | References |
| 1H NMR | Danio rerio (zebrafish) | 5 µm | 50 and 500 μg/L | 21 d | Intestine | Adult (5 months) | Changes in 36 metabolites. ↑ proline, propylene glycol, alanine, glutamine, leucine, ornithine, carnitine, threonine, TMAO; ↓ lysine, phenylalanine, tyrosine, linoleic acid, palmitic acid, triglycerides. | [31] |
| GC-MS | Danio rerio (zebrafish) | 100 nm | 250 and 2 × 104 items of PS MNPs in 50 mL | 72 hpf | Whole organism | Embryo | Changes in 508 metabolites. Disorders in unsaturated fatty acids, linoleic acid, taurine, hypotaurine, nicotinate, nicotinamide, alanine, aspartate, glutamate. | [22] |
| GC-MS | Danio rerio (zebrafish) | 5–50 μm | 100 and 1000 μg/L | 7 d | Whole organism | Embryo | Changes in 78 (5 μm) and 121 (50 μm) metabolites. Disorders in carbohydrates, fatty acids, amino acids, nucleic acids and others. | [114] |
| LC-MS/MS | Danio rerio (zebrafish) Perca fluviatilis (perch) | 5–12 µm | 1, 50 e 100 mg | 21 d | Gills and liver | Adult | Changes in 33 metabolites. Zebrafish gills: ↓ phenylalanine, carnitine, proline, salicylic and lactic acid, choline. Perch gills: ↑ phenylalanine, salicylic acid; ↓ acetyl-carnitine, alanine, glutamic and pyruvic acid. Zebrafish liver: ↑ adenine, adenosine, glutamine; ↓ hypoxanthine, uridine, deoxyadenosine, valine, arginine, phenylalanine, asparagine, proline. Perch liver: ↑ arginine, succinic acid, adenosine; ↓ hypoxanthine, oxoglutaric acid, citrulline, creatinine, adenine | [115] |
| LC-MS | Gobiocypris rarus (rare minnow) | 1 μm | 200 μg/L | 4 w | Liver | Subadult (3 months) | Changes in 41 metabolites. ↑ glyceraldehyde; cytosine, glucose, fructose, mannose; ↓ mannitol 1-phosphate, acetyl-phenylalanine, mannonate | [116] |
| UPLC-Q-TOF-MS | Oreochromis mossambicus (tilapia) | 100 nm | 20 mg/L and recovery | 7 d | Whole organism | Larvae | Changes in 203 metabolites. Disorders in fatty acyls, carboxylic acids and their derivatives, organooxygen compounds, keto acids and their derivatives. | [117] |
| LC-MS | Oreochromis niloticus (red tilapia) | 0.3, 5 and 70–90 μm | 100 μg/L | 14 d | Liver | Adult | Changes in 31 (0.3 μm), 40 (5 μm) and 23 (70–90 μm) metabolites. Disorders in amino acids, fatty acids, glycerophosphoethanolamines, glycerophosphoserines, glycerophosphocholines, purine nucleosides, eicosanoids. | [118] |
| 1H NMR | Oryzias javanicus (Javanese medaka) | 5 μm | 100, 500 and 1000 μg/L | 21 d | Gut | Adult | Changes in 9 metabolites. ↑ glucose, lactate, alanine, glutamate, glucoronate, valine, anserine, 2-hydroxyvalerate, creatine. | [119] |
| GC-MS | Oryzias melastigmas (marine medaka) | 10 and 200 μm | 10 mg/L | 60 d | Liver | Adult (8 months) | Changes in 83 metabolites. ↑ disaccharides, trisaccharides, fatty acids, fatty acid methyl and ethyl esters; ↓ monosaccharides, organic acids, amino acid. | [120] |
| LC-MS/MS | Sebastes Schlegelii (marine jacopever) | 5 μm and 100 nm | 0.23 mg/L | 15 d | Liver | Juvenile | Changes in 345 metabolites. Disorders in essential amino acids, omega-3 fatty acids, intermediate products of glucose metabolism and TCA intermediates. ↓ gluconic acid, cis-aconitate, malic acid, tyrosine, targinine, glycerol phospholipid | [121] |
| LC-MS | Xiphophorus helleri (swordtail fish) | 1 μm | 1 × 106 microspheres/L (B) and 1 × 107 microspheres/L (C) | 72 h | Liver | Adult (3 months) | Changes in 37 (B) and 103 (C) metabolites. ↑ 3-hydroxyanthranilic acid, histidine, citrulline, linoleic acid, pantothenate, xanthine. | [122] |
| AQUATIC INVERTEBRATES | ||||||||
| LC-MS/MS | Crassostrea gigas (Pacific oyster) | 6 and 50/60 μm | 1 × 104 particles/L | 14 d | Gills | Adult | Changes in 22 metabolites. ↑ asparagine, phenylalanine, glutathione, glucose-6-phosphate, carbohydrates, lactose, mannose; ↓ N-palmitoyl taurine, fatty acids. | [123] |
| 1H NMR | Daphnia magna (water flea) | 53 nm (PS-NH2), 62 nm (PS-COOH) | 3.2 μg/L | 37 d | Whole organism | Adult | Changes in 15 metabolites. ↑ alanine, asparagine, glutamate, glutamine, isoleucine, leucine, lysine, phenyl alanine, tyrosine, valine, lactate, methionine sulfoxide; ↓ glucose, glycogen, nucleic acids, isopropanol. | [124] |
| LC-MS/MS | Litopenaeus vannamei (whiteleg shrimp) | 2 μm | 0.02 to 1 mg/L | 72 h | Hepatopancreas | Post-larvae | Changes in 119 metabolites. ↑ amino acids and dipeptides, e.g., taurine, aspartic acid and alanine; ↓ glyceraldehyde and fatty acids, e.g., 3-methyladipic acid and leucinic acid. | [125] |
| 1H NMR | Mytilus galloprovincialis (Mediterranean mussel) | 3 μm | 50 particles/mL | 72 h | Gills and hepatopancreas | Changes in 10 (in gills) and 18 (in hepotopancreas) metabolites. Gills: ↑acetoacetate, ATP/ADP, mytilitol, betaine, taurine, homarine; ↓ alanine, glycine, succinate, acetylcholine. Hepatopancreas: ↑ isoleucine, leucine, valine, alanine, dimethylglycine, tyrosine, lactate, glycogen, glucose, betaine, taurine, homarine, glutathione; ↓ glycine, acetoacetate, succinate, malonate, hypotaurine. | [32,36] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eliso, M.C.; Billè, B.; Cappello, T.; Maisano, M. Polystyrene Micro- and Nanoplastics (PS MNPs): A Review of Recent Advances in the Use of -Omics in PS MNP Toxicity Studies on Aquatic Organisms. Fishes 2024, 9, 98. https://doi.org/10.3390/fishes9030098
Eliso MC, Billè B, Cappello T, Maisano M. Polystyrene Micro- and Nanoplastics (PS MNPs): A Review of Recent Advances in the Use of -Omics in PS MNP Toxicity Studies on Aquatic Organisms. Fishes. 2024; 9(3):98. https://doi.org/10.3390/fishes9030098
Chicago/Turabian StyleEliso, Maria Concetta, Barbara Billè, Tiziana Cappello, and Maria Maisano. 2024. "Polystyrene Micro- and Nanoplastics (PS MNPs): A Review of Recent Advances in the Use of -Omics in PS MNP Toxicity Studies on Aquatic Organisms" Fishes 9, no. 3: 98. https://doi.org/10.3390/fishes9030098