1. Introduction
Gilthead seabream (
Sparus aurata Linnaeus, 1758) is a highly valuable finfish for both fisheries and aquaculture industries in the Mediterranean Sea. Following FAO [
1], global capture and aquaculture production of this species in 2020 was approximately 8646 tonnes and 282,073 tonnes, respectively. It is a coastal euryhaline marine fish with a natural distribution that extends along the eastern Atlantic coasts and the Mediterranean Sea, up to the Black Sea [
2]. Previous studies have explored the natural genetic structuring of wild seabream populations across its entire distribution range using different genetic markers, including allozymes, mtDNA, microsatellites, and, recently, SNIPs (e.g., [
3,
4,
5,
6,
7,
8]). However, depending on the type and number of markers used, the existing literature on the genetic structuring of wild Gilthead seabreams has produced contradictory results with no evidence of simple isolation by distance among populations (e.g., [
3,
6,
9,
10]). Studies in both the Atlantic and Mediterranean Seas, at a variety of geographical scales, have indicated either an absence of genetic structure or a weak to strong population subdivision within and among different geographical areas (e.g., [
7,
8,
11,
12,
13,
14,
15,
16,
17,
18]). The long pelagic larval stage (up to 50 days at 17–18 °C in Gilthead seabreams [
2,
19]) is likely to contribute to migrant exchange among fish populations and seems to be one of the main reasons for the slight genetic differentiation and lack of geographic patterning among wild stocks in various fish species, including Gilthead seabreams (e.g., [
11,
12,
20,
21,
22,
23]). Additionally, accidental escapes of reared seabream, mainly due to technical or operational failures [
24,
25,
26], or the release of gametes into the wild through spawning within sea-cages [
27] may have impacted the genetic structure of local populations, potentially resulting in the introgression of foreign DNA into the wild gene pool. This hypothesis is further supported by the ability of reared escapees to survive in the natural environment for extended periods of time and to compete effectively for natural resources [
28,
29,
30].
Except for genetic markers, a variety of methods and approaches have been proposed to discriminate wild fish stocks (namely self-reproducing fish groups that are available for exploitation in a given area, with the members of each group having similar life history characteristics [
31]), including the analysis of phenotypic variability of different morphological traits of fish, such as body shape or otolith and scale morphology (e.g., [
32,
33,
34,
35,
36,
37]). Those traits are highly applicable in detecting differences in environmental conditions that fish experienced during their lifetime, particularly in the cases of marine species that breed in the open sea (e.g., Gilthead seabreams), where there are fewer barriers to dispersal and a small amount of gene flow between populations is sufficient to maintain the genetic homogeneity [
12,
20,
22,
38]. Previous studies have detected differences in body shape or in otolith and scale morphology between wild or between wild and reared Gilthead seabream populations of the western and eastern Mediterranean Sea [
39,
40], between fish of the wild and of a farmed origin in Greece [
41,
42,
43] as well as between wild, reared, and farm-associated seabream populations across the Adriatic Sea [
16,
44,
45,
46].
Otoliths are calcified structures found in the inner ears of teleosts and involved in hearing and balance systems [
47]. They consist of a calcium carbonate material that is deposited continuously on an organic matrix throughout the fish’s lifetime. Unlike scales and bones, otoliths are metabolically inert (i.e., once deposited, otolith material is unlikely to be resorbed or altered) and contain reliable fingerprints that provide information for the entire life cycle of the fish [
48,
49,
50]. The otolith shape results from the combined expression of its genotype and the conditions of the growing environment [
51,
52,
53,
54,
55,
56,
57,
58]. However, it also depends on the fish’s developmental stage, age, body size, sex, or sexual maturity status (e.g., [
50,
51,
59,
60,
61,
62,
63,
64]). At the individual level, bilateral differentiation in biomineralization processes induce morphological differences between the left and right pair of otoliths. At the population level, otolith fluctuating asymmetry (FA) refers to the random deviations from perfect bilateral symmetry induced by differences between the left and right body sides [
65]. Due to the high functional value of the otolith structures in fish balance and hearing [
66,
67], otolith FA is considered a sensitive indicator for examining the effects of environmental stress and/or environmental heterogeneity on the health status of wild fish populations (e.g., [
68,
69,
70,
71,
72]).
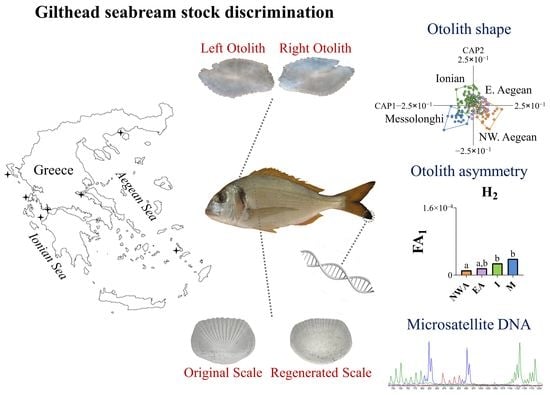
In the present study, we investigated the genetic population structure and spatial variation in the shape and asymmetry of otoliths in wild-caught Gilthead seabreams sourced from the Aegean and Ionian Seas (eastern Mediterranean). To control for the potential presence of aquaculture escapees among wild-caught individuals, we used the degree of scale regeneration (SRD, % of regenerated scales), which has been shown to be significantly higher in reared fish [
40,
41,
42,
73,
74], as an indicator to exclude potential aquaculture escapees (as previously demonstrated by [
41]) in the comparisons of otolith morphology (shape and asymmetry) and genetic composition between wild-caught fish of different geographical origins.
4. Discussion
In the present study, otolith shape and asymmetry analyses of wild-caught fish with a low possibility of being aquaculture escapees (L30 SRD group) revealed a phenotypic discrimination between northwestern Aegean and Ionian Gilthead seabream populations. However, a genetic analysis indicated an absence of genetic differentiation among the studied geographic fish groups. The otolith shape is a popular tool used for the discrimination among fish populations (e.g., [
53,
109,
110]) with significant value as an indicator of stock identity (e.g., [
47,
58]). The otolith shape has been shown to be affected by both biotic and abiotic environmental drivers during fish development, including water temperature [
60,
111,
112,
113], habitat depth [
114], type of substrate [
115], acidification levels [
116], food quantity [
117], and food composition [
55,
112]. Geladakis et al. [
35] suggested that observed phenotypic variability among wild fish stocks of the European sardine (
Sardina pilchardus Walbaum, 1792) could be attributed to differences in temperature and food availability between the Aegean and Ionian Seas. In Gilthead seabreams, the otolith shape was significantly different between distant wild populations of Greece and Spain [
40] or nearby populations with and without association with aquaculture farms in the Adriatic Sea [
46]. The lack of otolith shape differences between fish from the East Aegean and Ionian Seas in this study may potentially indicate that these two geographically distinct seabream populations have experienced common environmental conditions during critical developmental stages. It should be noted here that previous studies on stock discrimination using otolith shape (e.g., [
20,
118]) have demonstrated that fish sampling conducted over broad temporal scales with unbalanced sample sizes has the potential to influence the results obtained because the prevailing environmental conditions could affect otolith growth rates, thereby inducing otolith shape alterations [
51,
56,
119]. In this study, fish sampling was largely opportunistic, especially in the Ionian Sea, covering several years and different sampling sites and/or seasons. Unfortunately, with the exception of lagoon fisheries, such as the Messolonghi trap fishery, there is no other fishing practice in Greece targeting Gilthead seabreams at sea because the species has low abundance and is only caught occasionally as bycatch of bottom trawls and passive nets. Consequently, in order to ensure an adequate number of fish for the between-areas comparisons, we pooled all samples available from the broad geographic areas considered regardless of year, site, and season of collection. Sampling over broad temporal scales to acquire adequate sample sizes was also the approach followed in several other wild seabream population surveys in the past (e.g., December 2008–March 2009 [
17], July 2015–January 2017 [
18], October–December in 2015 and 2016 [
45,
46], July 2009–June 2010 [
15,
39,
40], 2001–2014 [
6]. Pooling data over large temporal and/or spatial regimes integrates temporal and/or areal heterogeneity in otolith shape, and the results of the analyses represent average conditions. However, in discriminating putative stocks, adequate and balanced sample sizes may not be possible in synoptic surveys for wild seabream. Thus, given no alternative, the pooling of data is a usual and accepted approach (e.g., [
6,
15,
17,
39,
40,
45,
46]). In an attempt to assess the effect on fish otolith shape of sampling at several occasions (different years and/or months), a PERMANOVA analysis was carried out for each of the four geographical areas considered, with a sampling survey (Year × Month combination) as a fixed factor (analyses not shown). PERMANOVA results and subsequent pair-wise comparisons showed that the effect of the sampling survey was only significant in the northwestern Aegean (for the right otolith) and the Ionian Sea (for both right and left otoliths). However, subsequent pairwise comparisons revealed that the samples that significantly differed in each of these two areas were those with a high proportion of fish with >60% regenerated scales (i.e., two samples: NW. Aegean, November 2015, 36.3% M60 fish; Ionian Sea, February 2018, 76.9% M60 fish [
Table S2]). This finding suggests that differences in otolith shape among samples collected in the same geographical area could be primarily attributed to the increased incidence of potential aquaculture escapees in certain samplings rather than a year and/or seasonal effect.
A genetic analysis revealed a higher genetic variation in wild compared to the reared Gilthead seabream populations. In agreement with our results, previous studies that assessed genetic variation using microsatellite markers found similar levels of observed and expected heterozygosity, mean number of alleles, and allelic richness in wild and reared seabream populations throughout Greece [
5,
14,
15,
120]. The absence of genetic structuring among the examined seabream samples indicates a genetic homogenization between the Aegean and Ionian Sea populations. Depending on the set of markers used, the existing literature suggests either an absence of genetic structure or a subtle genetic subdivision (F
ST = 0.009 [
5], F
ST = 0.002 [
14]). A recent geographically broad study by Maroso et al. [
6] within the Mediterranean Sea and Atlantic Ocean revealed a weak genetic subdivision (F
ST = 0.007) of wild Gilthead seabreams between the Aegean and Ionian Seas using 1159 genome-wide SNP markers. All these results combined with the absence of discernible physical or ecological barriers along with the long-lasting larval stage of Gilthead seabreams (~50 d [
12,
19]), enable high dispersal capacity, and support the exchange of individuals between populations of the Aegean and Ionian Seas, ultimately leading to a homogenized genetic pool.
In Greece, there has been an intensive rearing of Gilthead seabreams over the past three decades with rapid expansion in the previous decade [
14]. There is a continuous concern that this expansion of aquaculture activity has increased the likelihood of accidental escape events, which could potentially threaten the genetic status of wild fish populations and lead to unknown consequences for their fitness and survival. Reared fish may exhibit large genetic differences from adjacent wild populations mainly due to the different genetic background of broodstocks (i.e., different broodstock origins) or due to the application of selective breeding programs that aim to enhance specific traits [
5,
15,
23]. Our results (F
ST = 0.014–0.038) are in agreement with the existing literature, which has mainly shown weak genetic divergence between reared and wild Gilthead seabream populations along its distribution range, with F
STs ranging between 0.006–0.069 [
3,
5,
15,
18,
121,
122]. The genetic admixture of escapees with local wild populations could potentially introduce foreign DNA into the wild gene pool and damage local adaptations, but this stands for species with strong genetic structure like salmonids (F
ST = 0.034–0.2, [
123,
124,
125]). For such species, previous studies have shown that hybrids resulting from crosses between farmed and wild individuals could exhibit lower fitness (e.g., [
126]) or relative survival (e.g., [
127]) compared to native fish. However, following Sewall Wright’s guideline, F
ST-values between 0 and 0.05 indicate “no to little genetic differentiation”, F
ST-values between 0.05 and 0.15 represent moderate differentiation, F
ST-values between 0.15 and 0.25 are considered large, and F
ST-values above 0.25 show a very large degree of genetic differentiation [
128,
129,
130,
131,
132]. It is evident that this is not the case for Gilthead seabreams, which exhibit weak genetic structure with no signs of local adaptations in its populations, mainly due to the high dispersal capability of the species (a long-lived pelagic larval stage [
12,
19,
22]).
Scales have been utilized as a cost-effective and easily applicable morphological trait for identifying aquaculture escapees in the wild. In aquaculture facilities, friction and physical collisions among fish are intensified due to handling manipulations (e.g., counting, size-grading, and vaccination) and high population densities within sea-cages, leading to significantly high rates of scale loss [
73,
133,
134]. Lost scales are promptly replaced by new (regenerated scales), characterized by the presence of a large regeneration nucleus lacking growth
circulii [
74,
135,
136]. Based on our results on the otolith shape and asymmetry analyses, fish with a high degree of scale regeneration (31–60 and M60 SRD groups) exhibited increased otolith shape variability in comparison to the L30 group. Furthermore, fish with an M60 SRD exhibited significant differences in otolith shape and higher asymmetry levels compared to other SRD groups. In Gilthead seabreams, reared fish (>94% SRD) display distinct otolith shapes and higher asymmetry levels than wild fish [
41]. In addition, Geladakis et al. [
41] showed that wild-caught fish with SRD levels greater than 30% exhibit intermediate otolith phenotypes between wild fish with lower SRD (≤30%) and reared fish. Since scales are continuously lost during the rearing process [
74], we suggest the 60% SRD as a minimum threshold, beyond which wild-caught fish have an increased possibility of including individuals that escaped from net pens near harvest sizes, thus presenting phenotypes closely similar to those of reared fish. Following our results, a positive correlation emerged between SRD levels and the magnitude of otolith shape and asymmetry variation, suggesting that SRD can not only be used as a reliable index for detecting the presence of aquaculture escapees in the wild, but also to provide insights into the timing of fish escape from aquaculture net pens. On the other hand, a negative correlation was revealed between SRD levels and the genetic diversity (present study). Considering the lower levels of genetic diversity of the reared fish, this is in concordance with the proposal of Geladakis et al. [
41,
74] that the possibility of the presence of escapees increases as the level of SRD increases, as well. Given the utility of the SRD as a tool for identifying the presence of aquaculture escapees in the wild, the observed variations in SRD levels among wild-caught fish collected across the different years within each geographic group could potentially indicate a transient modification in the proportion of escaped individuals in the wild populations over time.
In our study, significant deviations from bilateral symmetry were observed in relation to both the origin within SRD groups and vice versa. These deviations were not restricted to only fluctuating asymmetry (FA), but also included directional asymmetry. Directional asymmetry (DA) refers to a consistent preference of a character toward greater development on one specific body side [
137]. In roundfish species, deviations from perfect bilateral symmetry in otolith morphology, expressed as FA or DA, have previously been observed at both early and later stages of fish development (e.g., [
41,
72,
112,
113,
138,
139,
140]. Otolith FA is regularly associated with different environmental stressors, and it is considered a sensitive indicator of fish health, performance, and population fitness [
65,
72]. On the other hand, otolith DA has been suggested to have a genetic origin and/or be a phenotypic plastic response induced by environmental drivers, such as water temperature, indicating a functional adaptation of fish hearing against environmental perturbations [
112,
113]. In the current work, a spatial variation was observed in the magnitude of otolith asymmetry (expressed as FA or DA) between different geographic groups. Depending on the descriptor examined, in fish with an L30 and 31–60 SRD, the group from Messolonghi lagoon exhibited significantly higher or lower FA or DA compared to other geographic groups. In fish with an L30 SRD, the Ionian group displayed significantly increased otolith shape DA (i.e., in H
2 descriptor) compared to the northwestern Aegean fish group. This finding is consistent with a previous study by Mahé et al. [
141], which also showed that the amplitude of otolith DA in bogue (
Boops boops Linnaeus, 1758) varied geographically, suggesting the potential use of directional bilateral asymmetry in otolith shape as a new method for stock discrimination. The observed shape differences in the right, but not in the left, otolith of the examined wild-caught individuals may be the underlying mechanism of the DA (the right side larger than the left side concerning the shape descriptor H
2) induced by early environmental conditions, such as temperature [
113]. Bilateral differentiation in the variability of otolith shape was also observed in Mahé et al. [
140], with significant discrimination between geographical stock units of bogue with respect to the shape of the right otolith but not the left otolith. However, an open question remains regarding the potential mechanism leading to the bilateral differentiation in otolith biomineralization processes during critical ontogenetic periods of fish and, therefore, the morphological differences between the left and right pair of otoliths.
During the late winter to early spring season (February to March), the Gilthead seabreams from the Ionian Sea temporarily enter the lagoon, which acts as a nursery ground, followed by migration to the open sea for breeding during October to December [
76]. The current study indicates that during this brief lagoon period of the Gilthead seabream life cycle, the environmental conditions substantially influence the morphology of their otoliths in terms of both the shape and asymmetry. Coastal lagoons are key ecosystems with increased natural productivity and intensive fishing exploitation by local communities [
75]. Fishing activity in these ecosystems is based on the seasonal entrance of young fish in the lagoons, where fish are then captured during their autumn-to-winter offshore migration [
76]. The Messolonghi–Etoliko lagoon complex is the largest in Greece and one of the most important coastal lagoon systems in the Mediterranean Sea [
142]. Because of the limited depth of the lagoons, meteorological changes rapidly affect the abiotic parameters of the water masses [
143]. The interaction between environmental conditions and genetic background could be the underlying mechanisms of otolith shape variability through determination on the relative growth of the different otolith parts (allometry) during fish growth [
53,
144]. Through the prism of otolith shape plasticity, a brief period of residence in lagoon ecosystems seems to significantly affect the otolith growth trajectories of seabream individuals and, therefore, the resulting otolith morphogenesis.