Deep Learning Applied to Intracranial Hemorrhage Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Deep Learning
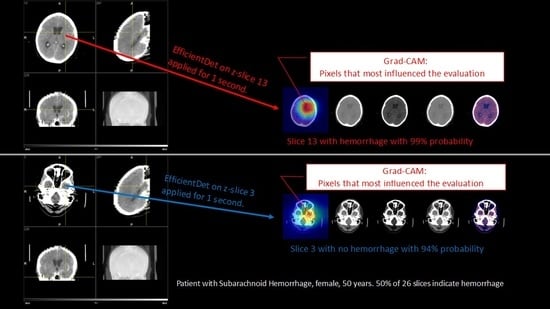
2.2. Grad-CAM
- Firstly, our model provides an output as to whether the patient has ICH or not. Technically, the output is the probability of having ICH. The model outputs an affirmative answer if such a probability is greater than a fixed threshold
- Our model further provides a color map on the input image where the red area corresponds to the pixels in the image which have been determinant in the decision.
2.3. Dataset
- Train: 97,525 with ICH, 580,934 without ICH.
- Validation: 5401 with ICH, 31,174 without ICH.
- Test: 5007 with ICH, 32,758 without ICH.
2.4. Metrics
2.5. Ethics
3. Results
3.1. Kaggle Test Dataset
3.2. Test Results Obtained with Clinical Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Some Technical Details
Appendix A.1. ResNet, EfficientDet and EffClass

- Upsampling approaches such as upsampling with nearest interpolation, lead to the same problem since we are not adding any information.
- Even without adding any information, ResNet would learn from these features, but, as ResNet receives (channels, height, width) as input, we would need an intermediate step, namely, to reduce 320 (5 sets of 64 filters each one) channels to only 3.


- (1)
- Load EfficientDet. We obtained a pre-trained EfficientDet-D0. In this step, we used the implementation from the timm library [38]. The timm library can be installed using Pypi.
- (2)
- Cut the regression and classification heads. We removed the regression and classification heads and only used the EfficientNet backbone and the BiFPN layers. Figure A4 shows this step and the previous one.
- (3)
- Deconvolution. Different deconvolution operations (transposed convolutions) were applied to the feature maps to get feature maps of as dimension. A total of 42 feature maps were passed to the convolution step. The number of channels returned by every deconvolution differed, depending on the resolution of the feature maps, as can be seen in Figure A5.
- (4)
- Convolution. In this step, the number of channels was reduced to 3. Two convolution layers were applied, as shown in Figure A6.
- (5)
- ResNet. We called a pre-trained ResNet18 from torchvision.models and cut the last layer as we did in the ResNet implementation section. The convolution layers´ output was passed to the ResNet as input.




Appendix A.2. Grad-CAM
- Step 1
- Compute the gradient of with respect to the feature map activations of a convolutional layer, i.e., , where is the raw output of the neural network for class c, before the softmax is applied to transform the raw score into a probability.
- Step 2
- Apply global average pooling to the gradients over the width dimension (indexed by i) and the height dimension (indexed by j) to obtain the neuron importance weights , producing k weights:
- Step 3
- Perform a weighted combination of the feature map activations with the weights, , calculated in the previous step:
Appendix A.3. EffClass Implementation
- Configuration directory. This directory contained the YAML files that governed the experiments. There were three types of configuration files corresponding to three different possible phases: training, validation, and testing.
- Experiments directory. If this directory did not exist, it was created automatically. It contained directories with the experimental code and, inside of them, there were loss and accuracy graphs, the configuration file used in the experiment, and the metrics (ROC-AUC, confusion matrix, and so on).
- Scripts directory. This directory contained the different scripts that were used in the different phases. The most important one was the config.py file, which led the execution and loaded the configuration from the YAML config file.
- Main file. The main.py file just received YAML config files and called the config.py and the corresponding task script, allowing us to execute multiple experiments.

References
- Feigin, V.L.; Abajour, A.A.; Abate, K.H.; Abd-Allah, F.; Abdulle, A.M.; Abera, S.F.; Abyu, G.Y.; Muktar, B.A.; Aichour, A.N.; Aichour, M.T.E.; et al. Global, regional, and national burden of neurological disorders during 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Neurol. 2017, 16, 877–897. [Google Scholar] [CrossRef]
- Caceres, J.A.; Goldstein, J.N. Intracranial hemorrhage. Emerg. Med. Clin. N. Am. 2012, 30, 771–794. [Google Scholar] [CrossRef]
- Fogelholm, R.; Murros, K.; Rissanen, A.; Avikainen, S. Long term survival after primary intracerebral haemorrhage: A retrospective population based study. J. Neurol. Neurosurg. Psychiatry 2005, 76, 1534–1538. [Google Scholar] [CrossRef]
- Jaja, B.N.; Cusimano, M.D.; Etminan, N.; Hanggi, D.; Hasan, D.; Ilodigwe, D.; Lantigua, H.; Le Roux, P.; Lo, B.; Louffat-Olivares, A.; et al. Clinical prediction models for aneurysmal subarachnoid hemorrhage: A systematic review. Neurocrit. Care 2013, 18, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Etminan, N.; Chang, H.S.; Hackenberg, K.; de Rooij, N.K.; Vergouwen, M.D.I.; Rinkel, G.J.E.; Algra, A. Worldwide Incidence of Aneurysmal Subarachnoid Hemorrhage According to Region, Time Period, Blood Pressure, and Smoking Prevalence in the Population: A Systematic Review and Meta-analysis. JAMA Neurol. 2019, 76, 588–597. [Google Scholar] [CrossRef]
- Al-Kawaz, M.N.; Hanley, D.F.; Ziai, W. Advances in Therapeutic Approaches for Spontaneous Intracerebral Hemorrhage. Neurotherapeutics 2020, 17, 1757–1767. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Fuad, M.T.H.; Fime, A.A.; Sikder, D.; Iftee, M.A.R.; Rabbi, J.; Al-Rakhami, M.S.; Gumaei, A.; Sen, O.; Fuad, M.; Islam, M.N. Recent Advances in Deep Learning Techniques for Face Recognition. IEEE Access 2021, 9, 99112–99142. [Google Scholar] [CrossRef]
- Hernandez-Olivan, C.; Beltran, J.R. Music Composition with Deep Learning: A Review. arXiv 2021, arXiv:2108.12290. [Google Scholar]
- Pan, X.; Wang, M.; Wu, L.; Li, L. Contrastive Learning for Many-to-many Multilingual Neural Machine Translation. arXiv 2021, arXiv:2105.09501. [Google Scholar]
- Shorfuzzaman, M.; Hossain, M.S. MetaCOVID: A Siamese neural network framework with contrastive loss for n-shot diagnosis of COVID-19 patients. Pattern Recognit. 2021, 113, 107700. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Zhou, T.; Ji, G.; Zhou, Y.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Inf-Net: Automatic COVID-19 Lung Infection Segmentation from CT Images. IEEE Trans. Med. Imaging 2020, 39, 2626–2637. [Google Scholar] [CrossRef] [PubMed]
- Nair, T.; Precup, D.; Arnold, D.L.; Arbel, T. Exploring uncertainty measures in deep networks for Multiple sclerosis lesion detection and segmentation. Med. Image Anal. 2020, 59, 1557. [Google Scholar] [CrossRef] [PubMed]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2019, 128, 336–359. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Tan, M.; Pang, R.; Le, Q.V. EfficientDet: Scalable and Efficient Object Detection. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 10778–10787. [Google Scholar] [CrossRef]
- Schwartz, R.; Dodge, J.; Smith, N.A.; Etzioni, O. Green AI. arXiv 2019, arXiv:1907.10597. [Google Scholar] [CrossRef]
- Burduja, M.; Ionescu, R.T.; Verga, N. Accurate and Efficient Intracranial Hemorrhage Detection and Subtype Classification in 3D CT Scans with Convolutional and Long Short-Term Memory Neural Networks. Sensors 2020, 20, 5611. [Google Scholar] [CrossRef]
- Gildenblat, J. Contributors. PyTorch Library for CAM Methods. 2021. Available online: https://github.com/jacobgil/pytorchgrad-cam (accessed on 16 November 2022).
- Kaggle Competition: RSNA intracranial Hemorrhage Detection. Available online: https://www.kaggle.com/c/rsna-intracranialhemorrhage-detection (accessed on 25 January 2021).
- Diagnostic Radiology Physics; Non-serial Publications; International Atomic Energy Agency: Vienna, Austria, 2014; p. 259.
- Voter, A.F.; Meram, E.; Garrett, J.W.; Yu, J.J. Diagnostic Accuracy and Failure Mode Analysis of a Deep Learning Algorithm for the Detection of Intracranial Hemorrhage. J. Am. Coll. Radiol. 2021, 18, 1143–1152. [Google Scholar] [CrossRef]
- Kuo, W.; Häne, C.; Mukherjee, P.; Malik, J.; Yuh, E.L. Expert-level detection of acute intracranial hemorrhage on head computed tomography using deep learning. Proc. Natl. Acad. Sci. USA 2019, 116, 22737–22745. [Google Scholar] [CrossRef]
- Kishan Das Menon, H.; Janardhan, V. Intracranial hemorrhage detection. In Materials Today: Proceedings, Proceedings of the International Conference on Nanoelectronics, Nanophotonics, Nanomaterials, Nanobioscience & Nanotechnology, Kottayam, India, 29–30 April 2021; IEEE: Hoboken, NJ, USA, 2021; Volume 43, pp. 3706–3714. [Google Scholar] [CrossRef]
- Sage, A.; Badura, P. Intracranial Hemorrhage Detection in Head CT Using Double-Branch Convolutional Neural Network, Support Vector Machine, and Random Forest. Appl. Sci. 2020, 10, 7577. [Google Scholar] [CrossRef]
- Gruschwitz, P.; Grunz, J.P.; Kuhl, P.J.; Kosmala, A.; Bley, T.A.; Petritsch, B.; Heidenreich, J.F. Performance testing of a novel deep learning algorithm for the detection of intracranial hemorrhage and first trial under clinical conditions. Neurosci. Inform. 2021, 1, 100005. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, J.S.; Kim, T.Y.; Kim, Y.S. Detection and classification of intracranial haemorrhage on CT images using a novel deep-learning algorithm. Sci. Rep. 2020, 10, 20546. [Google Scholar] [CrossRef]
- Kim, J.S.; Cho, Y.; Lim, T.H. Prediction of the Location of the Glottis in Laryngeal Images by Using a Novel Deep-Learning Algorithm. IEEE Access 2019, 7, 79545–79554. [Google Scholar] [CrossRef]
- AIDoc Medical Ltd. Available online: https://www.aidoc.com/ (accessed on 25 January 2021).
- Wang, X.; Shen, T.; Yang, S.; Lan, J.; Xu, Y.; Wang, M.; Zhang, J.; Han, X. A deep learning algorithm for automatic detection and classification of acute intracranial hemorrhages in head CT scans. NeuroImage Clin. 2021, 32, 2785. [Google Scholar] [CrossRef] [PubMed]
- Brinjikji, W.; Abbasi, M.; Arnold, C.; Benson, J.C.; Braksick, S.A.; Campeau, N.; Carr, C.M.; Cogswell, P.M.; Klaas, J.P.; Liebo, G.B.; et al. e-ASPECTS software improves interobserver agreement and accuracy of interpretation of aspects score. Intervent. Neuroradiol. 2021, 27, 1861. [Google Scholar] [CrossRef] [PubMed]
- Arbabshirani, M.R.; Fornwalt, B.K.; Mongelluzzo, G.J.; Suever, J.D.; Geise, B.D.; Patel, A.A.; Moore, G.J. Advanced machine learning in action: Identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. NPJ Digit. Med. 2018, 1, 9. [Google Scholar] [CrossRef]
- Brainscan.ai. Available online: https://brainscan.ai/ (accessed on 25 January 2021).
- Brzeski, A.; Gdańsk University of Technology, Gdańsk, Poland. Personal communication, 2019.
- Viniavskyi, O.; Dobko, M.; Dobosevych, O. Weakly-Supervised Segmentation for Disease Localization in Chest X-ray Images. In Proceedings of the Artificial Intelligence in Medicine; Michalowski, M., Moskovitch, R., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 249–259. [Google Scholar]
- Panwar, H.; Gupta, P.K.; Siddiqui, M.K.; Morales-Menendez, R.; Bhardwaj, P.; Singh, V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-Scan images. Chaos Solit. Fractt. 2020, 140, 110190. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Wightman, R. PyTorch Image Models. 2019. Available online: https://github.com/rwightman/pytorch-image-models (accessed on 16 November 2022).
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Advances in Neural Information Processing Systems 32; Wallach, H., Larochelle, H., Beygelzimer, A., d’Alché-Buc, F., Fox, E., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2019; pp. 8024–8035. [Google Scholar]


| Model (TH) | ACC | TPR | TNR | PPV | NPV | PR | ROC |
|---|---|---|---|---|---|---|---|
| EffClass (0.5) | 0.927 | 0.914 | 0.94 | 0.938 | 0.916 | 0.979 | 0.978 |
| EffClass (0.7) | 0.918 | 0.872 | 0.965 | 0.961 | 0.883 | 0.979 | 0.978 |
| EffClass (0.9) | 0.881 | 0.774 | 0.987 | 0.984 | 0.814 | 0.979 | 0.978 |
| Model (TH) | ACC | TPR | TNR | PPV | NPV | N | ICH (%) |
|---|---|---|---|---|---|---|---|
| Aidoc (FDA/501 k) [22] | 92.9% (*) | 93.6% (86.6–97.6) | 92.3% (85.4–96.6) | 91.7% (84.9–95.6) | 94.1% (88.4–97.2) | 198 | 47.5% |
| Aidoc [22] | 97.2% (*) | 92.3% (88.9–94.8) | 97.7% (97.2–98.2) | 81.3% (77.6–84.5) | 99.2% (98.8–99.4) | 3605 | 9.7% |
| EffClass (5%) | 95.5% (93.9–96.7) | 94.9% (92.1–96.9) | 95.8% (93.9–97.3) | 93.6% (90.8–95.6) | 96.7% (94.9–97.8) | 947 | 39.2% |
| EffClass (10%) | 93.6% (91.8-95.0) | 88.1% (84.4–91.3) | 97.0% (95.3–98.3) | 95.1% (92.3–96.9) | 92.7% (90.6–94.4) | 947 | 39.2% |
| EffClass test (2.5%) 55 patients | 94.6% (84.9–98.9) | 100.0% (92.5–100.0) | 62.5% (24.5–91.5) | 94.0% (86.5–97.5) | 100.0% (**) | 55 | 85.5% |
| EffClass test (5%) 55 patients | 96.4% (87.5–99.6) | 100% (92.5–100) | 75.0% (34.9–96.8) | 95.9% (87.6–98.7) | 100.0% (**) | 55 | 85.5% |
| EffClass test (10%) 55 patients | 100.0% (93.5–100.0) | 100.0% (92.5–100.0) | 100.0% (63.1–100.0) | 100.0% (**) | 100.0% (**) | 55 | 85.5% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cortés-Ferre, L.; Gutiérrez-Naranjo, M.A.; Egea-Guerrero, J.J.; Pérez-Sánchez, S.; Balcerzyk, M. Deep Learning Applied to Intracranial Hemorrhage Detection. J. Imaging 2023, 9, 37. https://doi.org/10.3390/jimaging9020037
Cortés-Ferre L, Gutiérrez-Naranjo MA, Egea-Guerrero JJ, Pérez-Sánchez S, Balcerzyk M. Deep Learning Applied to Intracranial Hemorrhage Detection. Journal of Imaging. 2023; 9(2):37. https://doi.org/10.3390/jimaging9020037
Chicago/Turabian StyleCortés-Ferre, Luis, Miguel Angel Gutiérrez-Naranjo, Juan José Egea-Guerrero, Soledad Pérez-Sánchez, and Marcin Balcerzyk. 2023. "Deep Learning Applied to Intracranial Hemorrhage Detection" Journal of Imaging 9, no. 2: 37. https://doi.org/10.3390/jimaging9020037