Transcriptome Analysis of Diploid and Autotetraploid Hemerocallis Response to Drought Stress
Abstract
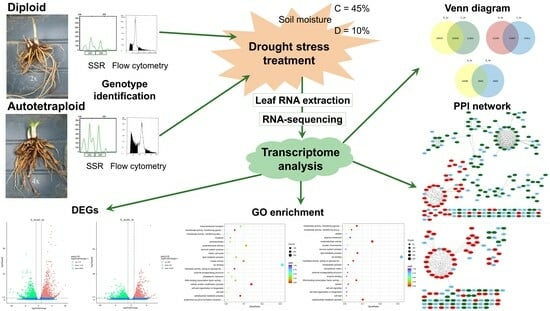
:1. Introduction
2. Materials and Methods
2.1. Plant Material, Cultivation, and Treatment
2.2. RNA Isolation, cDNA Library Preparation and Sequencing
2.3. De Novo Transcriptome Analysis
2.4. Functional Annotation of Unigenes
2.5. Gene Functional Annotation
2.6. Differentially Expressed Genes Venn Diagram, Volcano Plot, GO and KEGG Enrichment Analysis
2.7. Protein–Protein Interaction Analysis
3. Results
3.1. De Novo Transcriptome Sequencing and Quality Control of Bio-Project Data
3.2. Annotation of Unigenes in Hemerocallis spp. Transcriptome
3.3. Differentially Expressed Genes (DEGs) to Drought Stress Response in Diploid and Autotetraploid Daylily Leaves
3.4. Protein–Protein Interactions (PPI) Network during Drought Stress
4. Discussion
4.1. De Novo Transcriptome Sequencing of Daylily cv. Trahlyta
4.2. Ploidy Effect on DEGs and Pathways
4.3. Drought Resistance in Tetraploid Daylilies in PPIs Networks
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Takahashi, H.; Fukuhara, T.; Kitazawa, H.; Kormelink, R. Virus latency and the impact on plants. Front. Microbiol. 2019, 10, 2764. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Sarsaiya, S.; Wu, Q.; Lu, Y.; Shi, J. A review of plant leaf fungal diseases and its environment speciation. Bioengineered 2019, 10, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Safdar, H.; Amin, A.; Shafiq, Y.; Ali, A.; Yasin, R.; Shoukat, A.; Hussan, M.U.; Sarwar, M.I. A review: Impact of salinity on plant growth. Nat. Sci. 2019, 17, 34–40. [Google Scholar] [CrossRef]
- Sandeep, G.; Vijayalatha, K.R.; Anitha, T. Heavy metals and its impact in vegetable crops. Int. J. Chem. Stud. 2019, 7, 1612–1621. [Google Scholar]
- Kapoor, D.; Bhardwaj, S.; Landi, M.; Sharma, A.; Ramakrishnan, M.; Sharma, A. the impact of drought in plant metabolism: How to exploit tolerance mechanisms to increase crop production. Appl. Sci. 2020, 10, 5692. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Chen, S. Physiological and molecular mechanism involved in cold stress tolerance in plants. Plants 2020, 9, 560. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.U.; Chattha, M.U.; Khan, I.; Chattha, M.B.; Barbanti, L.; Aamer, M.; Iqbal, M.M.; Nawaz, M.; Mahmood, A.; Ali, A.; et al. Heat stress in cultivated plants: Nature, impact, mechanisms, and mitigation strategies—A review. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2021, 155, 211–234. [Google Scholar] [CrossRef]
- Jia, W.; Ma, M.; Chen, J.; Wu, S. Plant Morphological, Physiological and anatomical adaption to flooding stress and the underlying molecular mechanisms. Int. J. Mol. Sci. 2021, 22, 1088. [Google Scholar] [CrossRef]
- Skendžić, S.; Zovko, M.; Živković, I.P.; Lešić, V.; Lemić, D. The impact of climate change on agricultural insect pests. Insects 2021, 12, 440. [Google Scholar] [CrossRef]
- Sharma, A.; Abrahamian, P.; Carvalho, R.; Choudhary, M.; Paret, M.L.; Vallad, G.E.; Jones, J.B. Future of bacterial disease management in crop production. Annu. Rev. Phytopathol. 2022, 60, 259–282. [Google Scholar] [CrossRef]
- Karnieli, A.; Ohana-Levi, N.; Silver, M.; Paz-Kagan, T.; Panov, N.; Varghese, D.; Chrysoulakis, N.; Provenzale, A. Spatial and seasonal patterns in vegetation growth-limiting factors over Europe. Remote Sens. 2019, 11, 2406. [Google Scholar] [CrossRef]
- Zollinger, N.; Kjelgren, R.; Cerny-Koenig, T.; Kopp, K.; Koenig, R. Drought responses of six ornamental herbaceous perennials. Sci. Hortic. 2006, 109, 267–274. [Google Scholar] [CrossRef]
- Rafi, Z.N.; Kazemi, F.; Tehranifar, A. Effects of various irrigation regimes on water use efficiency and visual quality of some ornamental herbaceous plants in the field. Agric. Water Manag. 2019, 212, 78–87. [Google Scholar] [CrossRef]
- Toscano, S.; Ferrante, A.; Romano, D. Response of Mediterranean ornamental plants to drought stress. Horticulturae 2019, 5, 6. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Ashman, T.L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Hussain, H.A.; Hussain, S.; Khaliq, A.; Ashraf, U.; Anjum, S.A.; Men, S.; Wang, L. Chilling and drought stresses in crop plants: Implications, cross talk, and potential management opportunities. Front. Plant Sci. 2018, 9, 393. [Google Scholar] [CrossRef] [PubMed]
- Correia, S.; Braga, A.; Martins, J.; Correia, B.; Pinto, G.; Canhoto, J. Effects of polyploidy on physiological performance of acclimatized Solanum betaceum Cav. plants under water deficit. Forests 2023, 14, 208. [Google Scholar] [CrossRef]
- Ulum, F.B.; Hadacek, F.; Hörandl, E. Polyploidy improves photosynthesis regulation within the Ranunculus auricomus complex (Ranunculaceae). Biology 2021, 10, 811. [Google Scholar] [CrossRef]
- Tossi, V.E.; Martinez Tosar, L.J.; Laino, L.E.; Iannicelli, J.; Regalado, J.J.; Escandón, A.S.; Baroli, I.; Causin, H.F.; Pitta-Álvarez, S.I. Impact of polyploidy on plant tolerance to abiotic and biotic stresses. Front. Plant Sci. 2022, 13, 869423. [Google Scholar] [CrossRef]
- Hao, G.Y.; Lucero, M.E.; Sanderson, S.C.; Zacharias, E.H.; Holbrook, N.M. Polyploidy enhances the occupation of heterogeneous environments through hydraulic related trade-offs in Atriplex canescens (Chenopodiaceae). New Phytol. 2013, 197, 970–978. [Google Scholar] [CrossRef]
- Khalid, M.F.; Morillon, R.; Anjum, M.A.; Ejaz, S.; Rao, M.J.; Ahmad, S.; Hussain, S. Volkamer lemon tetraploid rootstock transmits the salt tolerance when grafted with diploid kinnow mandarin by strong antioxidant defense mechanism and efficient osmotic adjustment. J. Plant Growth Regul. 2022, 41, 1125–1137. [Google Scholar] [CrossRef]
- Barceló-Anguiano, M.; Holbrook, N.M.; Hormaza, J.I.; Losada, J.M. Changes in ploidy affect vascular allometry and hydraulic function in Mangifera indica trees. Plant J. 2021, 108, 541–554. [Google Scholar] [CrossRef]
- del Pozo, J.C.; Ramirez-Parra, E. Deciphering the molecular bases for drought tolerance in Arabidopsis autotetraploids. Plant Cell Environ. 2014, 37, 2722–2737. [Google Scholar] [CrossRef] [PubMed]
- Pomatto, E.; Larcher, F.; Caser, M.; Gaino, W.; Devecchi, M. Evaluation of different combinations of ornamental perennials for sustainable management in urban greening. Plants 2023, 12, 3293. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Sun, Y.; Wang, L.; Wang, J.; Wu, B.; Yan, T.; Jia, Y. An investigation of the anti-depressive properties of phenylpropanoids and flavonoids in Hemerocallis citrina Baroni. Molecules 2022, 27, 5809. [Google Scholar] [CrossRef] [PubMed]
- Mlcek, J.; Rop, O. Fresh edible flowers of ornamental plants—A new source of nutraceutical foods. Trends Food Sci. Technol. 2011, 22, 561–569. [Google Scholar] [CrossRef]
- Hao, Z.; Liang, L.; Liu, H.; Yan, Y.; Zhang, Y. Exploring the extraction methods of phenolic compounds in daylily (Hemerocallis citrina Baroni) and its antioxidant activity. Molecules 2022, 27, 2964. [Google Scholar] [CrossRef]
- Li, S.; Ji, F.; Hou, F.; Shi, Q.; Xing, G.; Chen, H.; Weng, Y.; Kang, X. Morphological, palynological and molecular assessment of Hemerocallis core collection. Sci. Hortic. 2021, 285, 110181. [Google Scholar] [CrossRef]
- Chung, M.G.; Kang, S.S. Morphometric analysis of the genus Hemerocallis L. (Liliaceae) in Korea. J. Plant Res. 1994, 107, 165–175. [Google Scholar] [CrossRef]
- American Hemerocallis Society Database AHS 2023. Available online: https://www.daylilydatabase.org/ (accessed on 15 September 2023).
- Misiukevičius, E. Polyploidy in daylily (Hemerocallis L.) breeding. Sodinink. Daržininkyste 2019, 38, 3–15. [Google Scholar]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef]
- Novikova, P.Y.; Hohmann, N.; Van de Peer, Y. Polyploid Arabidopsis species originated around recent glaciation maxima. Curr. Opin. Plant Biol. 2018, 42, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Podwyszyńska, M.; Gabryszewska, E.; Sochacki, D.; Jasiński, A. Histogenic identification by cytological analysis of colchicine-induced polyploids of Hemerocallis. Acta Hortic. 2011, 886, 245–249. [Google Scholar] [CrossRef]
- Li, Z.; Pinkham, L.; Campbell, N.F.; Espinosa, A.C.; Conev, R. Development of triploid daylily (Hemerocallis) germplasm by embryo rescue. Euphytica 2009, 169, 313–318. [Google Scholar] [CrossRef]
- Podwyszyńska, M.; Gabryszewska, E.; Dyki, B.; Stępowska, A.A.; Kowalski, A.; Jasiński, A. Phenotypic and genome size changes (variation) in synthetic tetraploids of daylily (Hemerocallis) in relation to their diploid counterparts. Euphytica 2015, 203, 1–16. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Liang, Z.; Jia, M.; Cao, D. Study on the indication of polyploid of Hemerocallis fulva by trifluridine. J. Shanxi Agric. Sci. 2018, 12, 1997–2000. [Google Scholar]
- Misiukevičius, E.; Stanys, V. Induction and analysis of polyploids in daylily (Hemerocallis L.) plants. Zemdirbyste-Agriculture 2022, 109, 373–382. [Google Scholar] [CrossRef]
- Cai, X.; Liu, J.; Zhao, F.; Wang, X. Transcriptome analysis of response strategy in Hemerocallis fulva under drought stress. Genes Genom. 2023, 45, 593–610. [Google Scholar] [CrossRef]
- Misiukevičius, E.; Frercks, B.; Šikšnianienė, J.B.; Kącki, Z.; Gębala, M.; Akulytė, P.; Trilikauskaitė, E.; Stanys, V. Assessing the genetic diversity of daylily germplasm using SSR markers: Implications for daylily breeding. Plants 2023, 12, 1752. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Stuart, J. Transcriptome analysis: Approaches and applications for gene expression. Transcr. Open Access 2023, 9, 143. [Google Scholar]
- Mažeikienė, I.; Juškytė, A.D.; Bendokas, V.; Stanys, V. De Novo Transcriptome analysis of R. nigrum cv. Aldoniai in response to blackcurrant reversion virus infection. Int. J. Mol. Sci. 2022, 23, 9560. [Google Scholar] [CrossRef] [PubMed]
- Muyle, A.; Marais, G.A.B.; Bačovský, V.; Hobza, R.; Lenormand, T. Dosage compensation evolution in plants: Theories, controversies and mechanisms. Philos. Trans. R. Soc. B 2022, 377, 20210222. [Google Scholar] [CrossRef]
- Yang, X.; Lu, M.; Wang, Y.; Wang, Y.; Liu, Z.; Chen, S. Response mechanism of plants to drought stress. Horticulturae 2021, 7, 50. [Google Scholar] [CrossRef]
- Wang, Z.; Li, G.; Sun, H.; Ma, L.; Guo, Y.; Zhao, Z.; Gao, H.; Mei, L. Effects of drought stress on photosynthesis and photosynthetic electron transport chain in young apple tree leaves. Biol. Open 2018, 7, bio035279. [Google Scholar] [CrossRef]
- Mahmood, T.; Khalid, S.; Abdullah, M.; Ahmed, Z.; Shah, M.K.N.; Ghafoor, A.; Du, X. Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells 2019, 9, 105. [Google Scholar] [CrossRef]
- Geng, D.; Shen, X.; Xie, Y.; Yang, Y.; Bian, R.; Gao, Y.; Li, P.; Sun, L.; Feng, H.; Ma, F.; et al. Regulation of phenylpropanoid biosynthesis by MdMYB88 and MdMYB124 contributes to pathogen and drought resistance in apple. Hortic. Res. 2020, 7, 102. [Google Scholar] [CrossRef]
- Iqbal, S.; Wang, X.; Mubeen, I.; Kamran, M.; Kanwal, I.; Díaz, G.A.; Abbas, A.; Parveen, A.; Atiq, M.N.; Alshaya, H.; et al. Phytohormones trigger drought tolerance in crop plants: Outlook and future perspectives. Front. Plant Sci. 2022, 12, 3378. [Google Scholar] [CrossRef]
- Takahashi, F.; Kuromori, T.; Sato, H.; Shinozaki, K. Regulatory Gene Networks in Drought Stress Responses and Resistance in Plants. In Survival Strategies in Extreme Cold and Desiccation. Advances in Experimental Medicine and Biology; Iwaya-Inoue, M., Sakurai, M., Uemura, M., Eds.; Springer: Singapore, 2018; Volume 1081, pp. 189–214. [Google Scholar] [CrossRef]
- Wang, L.; Lee, M.; Ye, B.; Yue, G.H. Genes, pathways and networks responding to drought stress in oil palm roots. Sci. Rep. 2020, 10, 21303. [Google Scholar] [CrossRef]
- Hasan, M.M.; Liu, X.D.; Waseem, M.; Guang-Qian, Y.; Alabdallah, N.M.; Jahan, M.S.; Fang, X.W. ABA activated SnRK2 kinases: An emerging role in plant growth and physiology. Plant Signal. Behav. 2022, 17, 2071024. [Google Scholar] [CrossRef]








| Ploidy | Sample | Raw Reads | Raw Bases, Gb | Clean Reads | Clean Bases, Gb | Q20 | GC pct, % |
|---|---|---|---|---|---|---|---|
| 2x | C | 20,677,729 (±1,491,722) | 6.2 (±0.4) | 19,834,903 (±2,072,326) | 6.0 (±0.6) | 96.80 (±0.74) | 45.78 (±0.64) |
| D | 28,653,149 (±2,942,249) | 8.6 (±0.9) | 27,341,411 (±3,482,860) | 8.2 (±1.0) | 96.31 (±0.06) | 44.73 (±0.92) | |
| 4x | C | 22,504,410 (±644,583) | 6.8 (±0.2) | 21,555,141 (±438,035) | 6.5 (±0.2) | 96.71 (±0.76) | 46.21 (±1.12) |
| D | 25,293,629 (±465,420) | 7.6 (±0.2) | 23,803,623 (±195,953) | 7.1 (±0.1) | 94.47 (±1.25) | 45.93 (±0.38) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Misiukevičius, E.; Mažeikienė, I.; Gossard, J.; Starkus, A.; Stanys, V. Transcriptome Analysis of Diploid and Autotetraploid Hemerocallis Response to Drought Stress. Horticulturae 2023, 9, 1194. https://doi.org/10.3390/horticulturae9111194
Misiukevičius E, Mažeikienė I, Gossard J, Starkus A, Stanys V. Transcriptome Analysis of Diploid and Autotetraploid Hemerocallis Response to Drought Stress. Horticulturae. 2023; 9(11):1194. https://doi.org/10.3390/horticulturae9111194
Chicago/Turabian StyleMisiukevičius, Edvinas, Ingrida Mažeikienė, James Gossard, Aurelijus Starkus, and Vidmantas Stanys. 2023. "Transcriptome Analysis of Diploid and Autotetraploid Hemerocallis Response to Drought Stress" Horticulturae 9, no. 11: 1194. https://doi.org/10.3390/horticulturae9111194