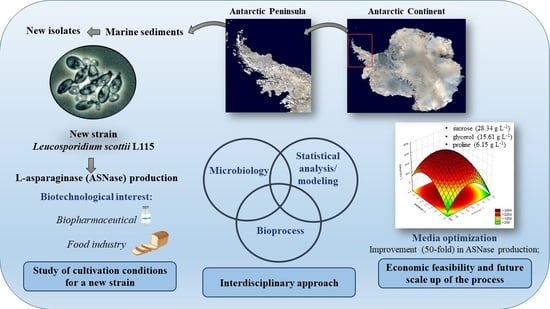
Selection and Optimization of Medium Components for the Efficient Production of L-Asparaginase by Leucosporidium scottii L115—A Psychrotolerant Yeast
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganism, Inoculum Preparation and Fermentation Conditions
2.1.1. Inoculum Preparation and Cultivation Conditions for the Plackett–Burman Design (PBD)
2.1.2. Inoculum Preparation and Cultivation Conditions for the Central Composite Design (CCD)
2.2. Statistical Analysis and Modeling
2.2.1. Plackett–Burman Design (PBD)
2.2.2. Central Composite Design (CCD)
2.3. Analyses
2.3.1. ASNase Activity Determination
2.3.2. Sucrose, Glycerol and Proline Quantification
3. Results and Discussion
3.1. Screening of Medium Components Using the PBD
3.2. Optimization of Medium Composition
| Yeast Strain | Medium Composition (g L−1) | ASNase (U L−1) | Reference |
|---|---|---|---|
| Different species including Rhodotorula, Candida utilis, Saccharomyces, Cryptococcus and Pichia (Hansenula) | Glucose, 20.0; L-asparagine, 5.0; K2HPO4, 1.0; KCl, 0.5; MgSO4·7H2O, 0.01; yeast extract, 1.0; pH 6.0 | from 5000 to 53,000 (depending on the strain) | [50] |
| Different species including Pichia (Hansenula), Cryptococcus, Sporobolomyces and Rhodotorula | Sucrose, 30.0; polypepton, 5.0; meat extract 5.0; yeast extract, 2.0; malt extract, 2.0; KH2PO4, 5.0; K2HPO4, 1.5; MgSO4·7H2O, 0.5 | from 111 to 1320 (depending on the strain) | [51] |
| S. cerevisiae X-2180-A2 | Growth medium: glucose, 20.0; yeast nitrogen base (without amino acids and ammonium sulfate), 2.0; and ammonium sulfate, 1.32 Production medium: glucose, 30.0; 20 mM potassium phosphate buffer, pH 7.0 | 3506 | [44] |
| Candida utilis | Glucose, 20.0; yeast nitrogen base (without amino acids and ammonium sulfate), 2.0; and peptone, 2.0 | 800 | [52] |
| Trichosporon asahii IBBLA1 | Glucose, 2; L-asparagine or glutamine or filter-sterilized urea, 10; KH2PO4, 1.52; KCl, 0.52; MgSO4·7H2O, 0.52; trace amounts of FeSO4·7H2O, ZnSO4·7H2O and CuNO3·3H2O | 20,570 | [6] |
| Rhodosporidium toruloides CBS14, ATCC10788, Rhodotorula glutinis NCYC59, ATCC90950, Rhodotorula rubra MTCC248 | Mannitol, 20.0; L-asparagine, 5.0; K2HPO4, 1.2; KH2PO4, 6.0; KCl, 0.5; MgSO4·7H2O, 0.01; yeast extract, 1.0; pH 6.0 | 583,000 | [53] |
| Leucosporidium scottii L 115 | Sucrose, 28.34; glycerol, 15.61; proline, 6.15; KCl, 0.52; MgSO4·7H2O, 0.52; CuNO3·3H2O, 0.001; ZnSO4·7H2O, 0.001; and FeSO4·7H2O, 0.001; pH 5.0 | 2850 | Present study |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Robinson, C.H. Cold adaptation in Arctic and Antarctic fungi. New Phytol. 2001, 151, 341–353. [Google Scholar] [CrossRef]
- Zhang, C.; Kim, S.-K. Research and Application of Marine Microbial Enzymes: Status and Prospects. Mar. Drugs 2010, 8, 1920–1934. [Google Scholar] [CrossRef] [PubMed]
- Zaky, A.S.; Tucker, G.A.; Daw, Z.Y.; Du, C. Marine yeast isolation and industrial application. FEMS Yeast Res. 2014, 14, 813–825. [Google Scholar] [CrossRef]
- Duarte, A.; Dayo-Owoyemi, I.; Nobre, F.S.; Pagnocca, F.C.; Chaud, L.C.S.; Pessoa, A.; Felipe, M.D.G.A.; Sette, L.D. Taxonomic assessment and enzymes production by yeasts isolated from marine and terrestrial Antarctic samples. Extremophiles 2013, 17, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Freire, R.K.B.; Mendonça, C.M.N.; Ferraro, R.B.; Moguel, I.S.; Tonso, A.; Lourenço, F.R.; Santos, J.H.P.L.; Sette, L.D.; Pessoa Jr, A. Glutaminase-free L-asparaginase production by Leucosporidium muscorum isolated from Antarctic marine-sediment. Prep. Biochem. Biotechnol. 2020, 51, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Ashok, A.; Doriya, K.; Rao, J.V.; Qureshi, A.; Tiwari, A.K.; Kumar, D.S. Microbes Producing L-Asparaginase free of Glutaminase and Urease isolated from Extreme Locations of Antarctic Soil and Moss. Sci. Rep. 2019, 9, 1423. [Google Scholar] [CrossRef]
- Muneer, F.; Siddique, M.H.; Azeem, F.; Rasul, I.; Muzammil, S.; Zubair, M.; Afzal, M.; Nadeem, H. Microbial l-asparaginase: Purification, characterization and applications. Arch. Microbiol. 2020, 202, 967–981. [Google Scholar] [CrossRef]
- Xu, F.; Oruna-Concha, M.-J.; Elmore, J.S. The use of asparaginase to reduce acrylamide levels in cooked food. Food Chem. 2016, 210, 163–171. [Google Scholar] [CrossRef]
- Zuo, S.; Zhang, T.; Jiang, B.; Mu, W. Reduction of acrylamide level through blanching with treatment by an extremely thermostable l-asparaginase during French fries processing. Extremophiles 2015, 19, 841–851. [Google Scholar] [CrossRef]
- World Health Organization. Model List of Essential Medicines, 21st List; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Brumano, L.P.; da Silva, F.V.S.; Costa-Silva, T.A.; Apolinário, A.C.; Santos, J.H.P.M.; Kleingesinds, E.K.; Monteiro, G.; Rangel-Yagui, C.D.O.; Benyahia, B.; Junior, A.P. Development of L-Asparaginase Biobetters: Current Research Status and Review of the Desirable Quality Profiles. Front. Bioeng. Biotechnol. 2019, 6, 212. [Google Scholar] [CrossRef]
- Merck. Elspar Asparaginase; Ovation Pharmaceuticals: Deerfield, IL, USA, 2000. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2002/aspamer080102LB.pdf (accessed on 27 July 2022).
- European Medicines Agency. Assessment Report Spectrila; European Medicines Agency: London, UK, 2015. Available online: https://www.ema.europa.eu/en/documents/assessment-report/spectrila-epar-public-assessment-report_en.pdf (accessed on 8 August 2022).
- European Medicines Agency. Assessment Report Oncaspar; European Medicines Agency: London, UK, 2016. Available online: https://www.ema.europa.eu/en/documents/assessment-report/oncaspar-epar-public-assessment-report_en.pdf (accessed on 8 August 2022).
- Medicines Evaluation Board. Public Assessmen Report Crisantaspase; European Medicines Agency: London, UK, 2015; Available online: https://file.wuxuwang.com/hma/NL_H_3194_001_PARSummary.pdf (accessed on 8 August 2022).
- Asparaginase (Erwinia chrysanthemi) (Recombinant)-rywn. Am. J. Health Pharm. 2021, 78, 1919–1921. [CrossRef] [PubMed]
- El-Naggar, N.E.-A.; Moawad, H.; El-Shweihy, N.M.; El-Ewasy, S.M. Optimization of Culture Conditions for Production of the Anti-Leukemic Glutaminase Free L-Asparaginase by Newly Isolated Streptomyces olivaceus NEAE-119 Using Response Surface Methodology. BioMed Res. Int. 2015, 2015, 627031. [Google Scholar] [CrossRef] [PubMed]
- Narta, U.K.; Kanwar, S.S.; Azmi, W. Pharmacological and clinical evaluation of l-asparaginase in the treatment of leukemia. Crit. Rev. Oncol. 2007, 61, 208–221. [Google Scholar] [CrossRef]
- Doriya, K.; Kumar, D.S. Isolation and screening of l-asparaginase free of glutaminase and urease from fungal sp. 3 Biotech 2016, 6, 239. [Google Scholar] [CrossRef] [PubMed]
- Muso-Cachumba, J.J.; Antunes, F.A.F.; Peres, G.F.D.; Brumano, L.; Santos, J.; Da Silva, S.S. Current applications and different approaches for microbial l-asparaginase production. Braz. J. Microbiol. 2016, 47, 77–85. [Google Scholar] [CrossRef]
- Liu, Z.; Feist, A.M.; Dragone, G.; Mussatto, S.I. Lipid and carotenoid production from wheat straw hydrolysates by different oleaginous yeasts. J. Clean. Prod. 2020, 249, 119308. [Google Scholar] [CrossRef]
- Dinarvand, M.; Rezaee, M.; Masomian, M.; Jazayeri, S.D.; Zareian, M.; Abbasi, S.; Ariff, A.B. Effect of C/N Ratio and Media Optimization through Response Surface Methodology on Simultaneous Productions of Intra- and Extracellular Inulinase and Invertase from Aspergillus niger ATCC 20611. BioMed Res. Int. 2013, 2013, 508968. [Google Scholar] [CrossRef]
- Mussatto, S.I.; Roberto, I.C. Establishment of the optimum initial xylose concentration and nutritional supplementation of brewer’s spent grain hydrolysate for xylitol production by Candida guilliermondii. Process. Biochem. 2008, 43, 540–546. [Google Scholar] [CrossRef]
- Mussatto, S.I.; Fernandes, M.; Dragone, G.; Mancilha, I.M.; Roberto, I.C. Brewer’s spent grain as raw material for lactic acid production by Lactobacillus delbrueckii. Biotechnol. Lett. 2007, 29, 1973–1976. [Google Scholar] [CrossRef]
- Gancedo, J.M. Yeast Carbon Catabolite Repression. Microbiol. Mol. Biol. Rev. 1998, 62, 334–361. [Google Scholar] [CrossRef]
- Sanchez, S.; Demain, A.L. Metabolic regulation and overproduction of primary metabolites. Microb. Biotechnol. 2008, 1, 283–319. [Google Scholar] [CrossRef] [PubMed]
- Magasanik, B.; Kaiser, C.A. Nitrogen regulation in Saccharomyces cerevisiae. Gene 2002, 290, 1–18. [Google Scholar] [CrossRef]
- Margesin, R.; Schinner, F. Properties of cold-adapted microorganisms and their potential role in biotechnology. J. Biotechnol. 1994, 33, 1–14. [Google Scholar] [CrossRef]
- Segal-Kischinevzky, C.; Romero-Aguilar, L.; Alcaraz, L.D.; López-Ortiz, G.; Martínez-Castillo, B.; Torres-Ramírez, N.; Sandoval, G.; González, J. Yeasts Inhabiting Extreme Environments and Their Biotechnological Applications. Microorganisms 2022, 10, 794. [Google Scholar] [CrossRef]
- Moguel, I.S.; Yamakawa, C.K.; Pessoa, A., Jr.; Mussatto, S.I. L-asparaginase Production by Leucosporidium scottii in a Bench-Scale Bioreactor with Co-production of Lipids. Front. Bioeng. Biotechnol. 2020, 8, 576511. [Google Scholar] [CrossRef]
- Granato, D.; Calado, V.M.D.A. The use and importance of design of experiments (DOE) in process modelling in food science and technology. In Mathematical and Statistical Methods in Food Science and Technology; John Wiley & Sons, Ltd.: Chichester, UK, 2013; pp. 1–18. [Google Scholar] [CrossRef]
- Hymavathi, M.; Sathish, T.; Brahmaiah, P.; Prakasham, R.S. Impact of carbon and nitrogen sources on L-asparaginase pro-duction by isolated Bacillus circulans (MTCC 8574): Application of saturated Plackett-Burman Design. Chem. Biochem. Eng. Q. 2010, 24, 473–480. [Google Scholar] [CrossRef]
- Analytical Methods Committee. Experimental design and optimisation (4): Plackett–Burman designs. Anal. Methods 2013, 5, 1901–1903. [Google Scholar] [CrossRef]
- Freitas, M.; Souza, P.; Cardoso, S.; Cruvinel, K.; Abrunhosa, L.S.; Filho, E.X.F.; Inácio, J.; Pinho, D.B.; Pessoa, A.; Magalhães, P.O. Filamentous Fungi Producing l-Asparaginase with Low Glutaminase Activity Isolated from Brazilian Savanna Soil. Pharmaceutics 2021, 13, 1268. [Google Scholar] [CrossRef]
- Gulati, R.; Saxena, R.K.; Gupta, R. A rapid plate assay for screening l -asparaginase producing micro-organisms. Lett. Appl. Microbiol. 1997, 24, 23–26. [Google Scholar] [CrossRef]
- Narayana, K.J.P.; Kumar, K.G.; Vijayalakshmi, M. L-asparaginase production by Streptomyces albidoflavus. Indian J. Microbiol. 2008, 48, 331–336. [Google Scholar] [CrossRef]
- Bon, E.P.S.; Carvajal, E.; Stanbrough, M.; Rowen, D.; Magasanik, B. Asparaginase II of Saccharomyces cerevisiae. In Biotechnology for Fuels and Chemicals; Applied Biochemistry and Biotechnology Book Series; Davison, B.H., Wyman, C.E., Finkelstein, M., Eds.; Humana Press: Totowa, NJ, USA, 1997; Volume 63–65, pp. 203–212. [Google Scholar] [CrossRef]
- Godard, P.; Urrestarazu, A.; Vissers, S.; Kontos, K.; Bontempi, G.; van Helden, J.; André, B. Effect of 21 Different Nitrogen Sources on Global Gene Expression in the Yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2007, 27, 3065–3086. [Google Scholar] [CrossRef] [PubMed]
- Kaul, S.; Sharma, S.S.; Mehta, I.K. Free radical scavenging potential of L-proline: Evidence from in vitro assays. Amino Acids 2008, 34, 315–320. [Google Scholar] [CrossRef]
- Chen, X.-C.; Bai, J.-X.; Cao, J.-M.; Li, Z.-J.; Xiong, J.; Zhang, L.; Hong, Y.; Ying, H.-J. Medium optimization for the production of cyclic adenosine 3′,5′-monophosphate by Microbacterium sp. no. 205 using response surface methodology. Bioresour. Technol. 2009, 100, 919–924. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H. Proline as a stress protectant in yeast: Physiological functions, metabolic regulations, and biotechnological applications. Appl. Microbiol. Biotechnol. 2008, 81, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Posch, A.E.; Spadiut, O.; Herwig, C. Switching industrial production processes from complex to defined media: Method development and case study using the example of Penicillium chrysogenum. Microb. Cell Fact. 2012, 11, 88. [Google Scholar] [CrossRef]
- da Silva, G.P.; Mack, M.; Contiero, J. Glycerol: A promising and abundant carbon source for industrial microbiology. Biotechnol. Adv. 2009, 27, 30–39. [Google Scholar] [CrossRef]
- Dunlop, P.; Meyer, G.; Ban, D.; Roon, R. Characterization of two forms of asparaginase in Saccharomyces cerevisiae. J. Biol. Chem. 1978, 253, 1297–1304. Available online: https://www.jbc.org/content/253/4/1297.full.pdf (accessed on 8 August 2022). [CrossRef]
- Baskar, G.; Renganathan, S. Statistical and evolutionary optimisation of operating conditions for enhanced production of fungal l-asparaginase. Chem. Pap. 2011, 65, 798–804. [Google Scholar] [CrossRef]
- Farag, A.M.; Hassan, S.W.; Beltagy, E.A.; El-Shenawy, M.A. Optimization of production of anti-tumor l-asparaginase by free and immobilized marine Aspergillus terreus. Egypt. J. Aquat. Res. 2015, 41, 295–302. [Google Scholar] [CrossRef]
- Saxena, R.K.; Sinha, U. L-Asparaginase and glutaminase activities in the culture filtrates of Aspergillus nidulans. Curr. Sci. 1981, 50, 218–219. [Google Scholar]
- Souza, P.M.; de Freitas, M.M.; Cardoso, S.L.; Pessoa, A.; Guerra, E.N.S.; Magalhães, P.O. Optimization and purification of l-asparaginase from fungi: A systematic review. Crit. Rev. Oncol. Hematol. 2017, 120, 194–202. [Google Scholar] [CrossRef]
- Moguel, I.S. Production of L-Asparaginase of Pharmaceutical Nterest from Yeasts Isolated from the Antarctic continent. Ph.D. Thesis, University of São Paulo, São Paulo, Brazil, 2018. [Google Scholar] [CrossRef]
- Arima, K.; Sakamoto, T.; Araki, C.; Tamura, G. Production of Extracellularl-Asparaginases by Microorganisms. Agric. Biol. Chem. 1972, 36, 356–361. [Google Scholar] [CrossRef]
- Imada, A.; Igarasi, S.; Nakahama, K.; Isono, M. Asparaginase and Glutaminase Activities of Micro-organisms. J. Gen. Microbiol. 1973, 76, 85–99. [Google Scholar] [CrossRef]
- Kil, J.-O.; Kim, G.-N.; Park, I. Extraction of Extracellular L-Asparaginase from Candida utilis. Biosci. Biotechnol. Biochem. 1995, 59, 749–750. [Google Scholar] [CrossRef]
- Ramakrishnan, M.S.; Joseph, R. Characterization of an extracellular asparaginase of Rhodosporidium toruloides CBS14 exhibiting unique physicochemical properties. Can. J. Microbiol. 1996, 42, 316–325. [Google Scholar] [CrossRef]
- Vidya, J.; Vasudevan, U.M.; Soccol, C.R.; Pandey, A. Cloning, functional expression and characterization of L-asparaginase II from E. coli MTCC 739. Food Technol. Biotechnol. 2011, 49, 286–290. [Google Scholar]
- Girão, L.F.D.C.; da Rocha, S.L.G.; Sobral, R.S.; Bom, A.P.D.A.; Sampaio, A.L.F.; da Silva, J.G., Jr.; Ferrara, M.A.; Bon, E.P.D.S.; Perales, J. Saccharomyces cerevisiae asparaginase II, a potential antileukemic drug: Purification and characterization of the enzyme expressed in Pichia pastoris. Protein Expr. Purif. 2016, 120, 118–125. [Google Scholar] [CrossRef]
- Bjelic, S.; Brandsdal, B.O.; Åqvist, J. Cold Adaptation of Enzyme Reaction Rates. Biochemistry 2008, 47, 10049–10057. [Google Scholar] [CrossRef] [PubMed]


| Code | Variables | +1 (g L−1) | −1 (g L−1) |
|---|---|---|---|
| Nutrients considered as nitrogen source | |||
| A | Proline | 5 | 0 |
| B | Urea | 5 | 0 |
| C | L-Aspartate | 5 | 0 |
| D | Ammonium chloride | 5 | 0 |
| E | L-Arginine | 5 | 0 |
| F | L-Glutamine | 5 | 0 |
| G | L-Asparagine | 5 | 0 |
| H | L-Glutamate | 5 | 0 |
| Nutrients considered as carbon source | |||
| I | Maltose | 10 | 0 |
| J | Glycerol | 10 | 0 |
| K | Sucrose | 10 | 0 |
| L | Citric acid | 10 | 0 |
| Complex sources | |||
| M | Yeast extract | 2.5 | 0 |
| N | Soybean peptone | 2.5 | 0 |
| O | Corn steep solids | 2.5 | 0 |
| P | Potato dextrose broth | 2.5 | 0 |
| Dummy variables * | |||
| Q | D1 | 0 | 0 |
| R | D2 | 0 | 0 |
| S | D3 | 0 | 0 |
| R | D2 | 0 | 0 |
| Run | Variables * | ASNase (U L−1) | DCW (g L−1) | ASNase | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | (U gdcw−1) | |||
| 1 | - | + | - | - | - | + | + | - | + | + | + | - | + | + | - | - | + | + | - | 166.6 | 5.96 | 28.0 |
| 2 | + | - | + | - | - | - | + | + | + | + | - | - | - | + | + | - | - | + | + | 141.6 | 6.18 | 22.9 |
| 3 | - | + | - | - | - | - | - | + | + | + | + | + | + | - | + | + | - | - | + | 206.0 | 7.04 | 29.3 |
| 4 | + | - | + | - | + | + | - | + | - | + | + | - | + | + | - | + | - | - | - | 181.8 | 10.36 | 17.5 |
| 5 | + | + | - | + | - | + | + | + | - | - | + | + | - | + | + | - | - | - | - | 212.0 | 4.23 | 50.1 |
| 6 | + | + | + | - | + | - | + | - | + | - | + | - | - | - | + | + | + | - | - | 152.7 | 5.97 | 25.6 |
| 7 | + | + | + | + | - | + | - | - | + | + | - | + | - | - | - | + | - | + | - | 161.9 | 3.64 | 44.5 |
| 8 | - | + | + | - | + | + | + | + | - | + | - | + | - | - | - | - | + | - | + | 73.1 | 1.29 | 56.7 |
| 9 | - | - | + | + | + | - | + | + | + | - | + | + | + | - | - | - | - | + | - | 119.5 | 5.37 | 22.3 |
| 10 | + | - | - | + | + | - | - | - | + | + | + | + | - | + | - | - | + | - | + | 296.8 | 11.33 | 26.2 |
| 11 | + | + | - | + | + | - | - | + | - | + | - | - | + | - | + | - | + | + | - | 117.8 | 3.24 | 36.4 |
| 12 | - | + | + | + | - | - | - | + | - | - | + | - | - | + | - | + | + | + | + | 165.7 | 3.73 | 44.4 |
| 13 | + | - | + | - | - | + | - | - | - | - | + | + | + | - | + | - | + | + | + | 180.6 | 6.98 | 25.9 |
| 14 | + | + | - | - | + | - | + | - | - | - | - | + | + | + | - | + | - | + | + | 40.05 | 2.50 | 16.0 |
| 15 | - | + | + | + | + | + | - | - | + | - | - | - | + | + | + | - | - | - | + | 90.01 | 1.89 | 47.6 |
| 16 | - | - | + | + | - | - | + | - | - | + | - | + | + | + | + | + | + | - | - | 173.1 | 6.24 | 27.7 |
| 17 | - | - | - | - | + | + | - | + | + | - | - | + | - | + | + | + | + | + | - | 101.0 | 2.94 | 34.4 |
| 18 | - | - | - | + | + | + | + | - | - | + | + | - | - | - | + | + | - | + | + | 234.4 | 11.23 | 20.9 |
| 19 | + | - | - | + | - | + | + | + | + | - | - | - | + | - | - | + | + | - | + | 70.25 | 2.98 | 23.6 |
| 20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 39.77 | 1.07 | 37.2 |
| ASNase Production | DCW Production | |||||||
|---|---|---|---|---|---|---|---|---|
| Effect | S.E. | t Student | p-Value | Effect | S.E. | t Student | p-Value | |
| Intercept | 146.27 | 5.82 | 50.21 | 0.00001 | 5.20 | 0.31 | 33.77 | 0.00005 |
| Yeast extract | −33.39 | 5.82 | −5.73 | 0.010 * | −0.07 | 0.31 | −0.21 | 0.841 |
| Soybean peptone | 11.20 | 5.82 | 1.92 | 0.150 | 0.49 | 0.31 | 1.59 | 0.209 |
| Corn steep solids | 19.33 | 5.82 | 3.31 | 0.045 * | 0.60 | 0.31 | 1.96 | 0.144 |
| Potato dextrose broth | 4.91 | 5.82 | 0.84 | 0.461 | 0.90 | 0.31 | 2.94 | 0.060 * |
| L-Glutamine | −8.17 | 5.82 | −1.40 | 0.255 | −0.28 | 0.31 | −0.91 | 0.429 |
| L-Asparagine | −15.79 | 5.82 | −2.71 | 0.073 * | −0.02 | 0.31 | −0.08 | 0.939 |
| Maltose | −1.24 | 5.82 | −0.21 | 0.845 | 0.08 | 0.31 | 0.26 | 0.808 |
| Glycerol | 58.18 | 5.82 | 9.99 | 0.002 * | 2.89 | 0.31 | 9.36 | 0.002 * |
| L-Glutamate | −14.71 | 5.82 | −2.52 | 0.085 * | −0.94 | 0.31 | −3.06 | 0.054 |
| Sucrose | 90.78 | 5.82 | 15.58 | 0.001 * | 4.02 | 0.31 | 13.04 | 0.001 * |
| Proline | 18.63 | 5.82 | 3.20 | 0.049 * | 1.06 | 0.31 | 3.46 | 0.040 * |
| Urea | −25.31 | 5.82 | −4.34 | 0.022 * | −2.68 | 0.31 | −8.70 | 0.003 * |
| L-Aspartate | 14.51 | 5.82 | −2.49 | 0.088 * | −0.25 | 0.31 | −0.80 | 0.478 |
| Citric acid | 20.33 | 5.82 | 3.49 | 0.039 * | −0.10 | 0.31 | −0.34 | 0.755 |
| L-Arginine | −21.08 | 5.82 | −3.62 | 0.036* | 0.64 | 0.31 | 2.08 | 0.128 |
| Ammonium chloride | 25.76 | 5.82 | 4.42 | 0.021 * | 0.19 | 0.31 | 0.63 | 0.570 |
| Run | Proline (g L−1) | Sucrose (g L−1) | Glycerol (g L−1) | ASNase (U L−1) | |
|---|---|---|---|---|---|
| Experimental | Predicted | ||||
| 1 | 4 | 10 | 10 | 2489.46 | 2283.89 |
| 2 | 4 | 10 | 40 | 2563.13 | 2254.16 |
| 3 | 4 | 40 | 10 | 3012.48 | 2955.93 |
| 4 | 4 | 40 | 40 | 2328.63 | 2367.27 |
| 5 | 10 | 10 | 10 | 2110.09 | 1879.50 |
| 6 | 10 | 10 | 40 | 1939.43 | 1812.33 |
| 7 | 10 | 40 | 10 | 2048.70 | 2169.23 |
| 8 | 10 | 40 | 40 | 1535.50 | 1543.12 |
| 9 | 2 | 25 | 25 | 2081.85 | 2290.97 |
| 10 | 12 | 25 | 25 | 1259.26 | 1267.19 |
| 11 | 7 | 0 | 25 | 1456.93 | 1856.54 |
| 12 | 7 | 50 | 25 | 2374.05 | 2192.24 |
| 13 | 7 | 25 | 0 | 3108.25 | 3212.95 |
| 14 | 7 | 25 | 50 | 2556.99 | 2666.41 |
| 15 | 7 | 25 | 25 | 3202.20 | 3366.92 |
| 16 | 7 | 25 | 25 | 3382.46 | 3366.92 |
| 17 | 7 | 25 | 25 | 3251.87 | 3366.92 |
| Source | S0053\A | d.f. | MS | F-Value | p-Value |
|---|---|---|---|---|---|
| Proline (L) | 1,251,591 | 1 | 1,251,591 | 144.37 | 0.0068 * |
| Proline (Q) | 3,141,439 | 1 | 3,141,439 | 362.37 | 0.0027 * |
| Sucrose (L) | 137,444 | 1 | 137,444 | 15.85 | 0.0576 * |
| Sucrose (Q) | 2,195,471 | 1 | 2,195,471 | 253.25 | 0.0039 * |
| Glycerol (L) | 360,911 | 1 | 360,911 | 41.63 | 0.0231 * |
| Glycerol (Q) | 154,271 | 1 | 154,271 | 17.79 | 0.0518 * |
| Proline × Sucrose | 70,724 | 1 | 70,724 | 8.15 | 0.1038 |
| Proline × Glycerol | 679 | 1 | 679 | 0.07 | 0.8059 |
| Sucrose × Glycerol | 151,267 | 1 | 15,267 | 17.44 | 0.0528 * |
| Lack of fit | 452,698 | 5 | 90,540 | 10.44 | 0.1087 |
| Pure error | 17,338 | 2 | 8669 | ||
| Total SS | 6,734,472 | 16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moguel, I.S.; Yamakawa, C.K.; Brumano, L.P.; Pessoa, A., Jr.; Mussatto, S.I. Selection and Optimization of Medium Components for the Efficient Production of L-Asparaginase by Leucosporidium scottii L115—A Psychrotolerant Yeast. Fermentation 2022, 8, 398. https://doi.org/10.3390/fermentation8080398
Moguel IS, Yamakawa CK, Brumano LP, Pessoa A Jr., Mussatto SI. Selection and Optimization of Medium Components for the Efficient Production of L-Asparaginase by Leucosporidium scottii L115—A Psychrotolerant Yeast. Fermentation. 2022; 8(8):398. https://doi.org/10.3390/fermentation8080398
Chicago/Turabian StyleMoguel, Ignacio S., Celina K. Yamakawa, Larissa P. Brumano, Adalberto Pessoa, Jr., and Solange I. Mussatto. 2022. "Selection and Optimization of Medium Components for the Efficient Production of L-Asparaginase by Leucosporidium scottii L115—A Psychrotolerant Yeast" Fermentation 8, no. 8: 398. https://doi.org/10.3390/fermentation8080398