Staphylococcus aureus Small RNAs Possess Dephospho-CoA 5′-Caps, but No CoAlation Marks
Abstract
:1. Introduction
2. Results
2.1. Synthesis and Purification of dpCoA-RNA
2.2. Detection and Quantification of CoA and dpCoA-RNA
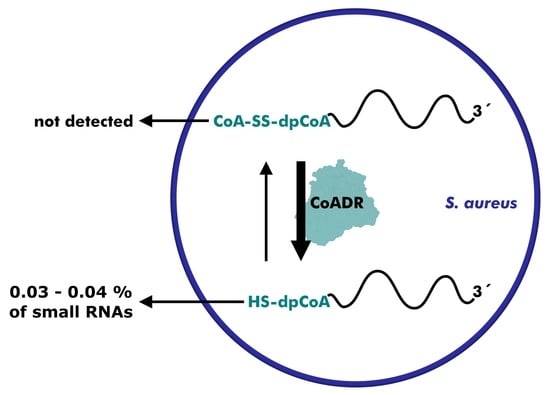
2.3. dpCoA Capping and CoAlation in S. aureus Small RNA Isolates
2.4. Reduction of dpCoA-RNA Disulfides by S. aureus CoADR
3. Discussion
4. Materials and Methods
4.1. General Procedures
4.2. Synthesis of 4-(Acrylamido)Phenylmercuric Chloride (APM)
4.3. Synthesis of 3-(Acrylamido)Phenylboronic Acid (APB)
4.4. Synthesis of 2-Mercaptopyridine-Activated Disulfides
4.5. Polyacrylamide Electrophoresis (PAGE)
4.6. Affinity Gel Electrophoresis (APM-PAGE and ABP-PAGE)
4.7. Synthesis of dpCoA-RNA
4.8. Synthesis of dpCoA-RNA Disulfides
4.9. Bacterial Growth
4.10. Molecular Cloning of S. aureus Coenzyme A Disulfide Reductase
4.11. Protein Expression
4.12. Small RNA Isolation
4.13. Quantification of CoA and dpCoA-RNA
4.14. S. aureus CoADR Reaction
4.15. Fluorescent Labeling of dpCoA-RNA
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fahey, R.C. Glutathione analogs in prokaryotes. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3182–3198. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.G.; Holmgren, A.; Arnér, E.; Schmidt, E.E. NADPH-dependent and -independent disulfide reductase systems. Free Radic. Biol. Med. 2018, 127, 248–261. [Google Scholar] [CrossRef]
- Newton, G.L.; Fahey, R.C.; Rawat, M. Detoxification of toxins by bacillithiol in Staphylococcus aureus. Microbiology 2012, 158, 1117–1126. [Google Scholar] [CrossRef] [Green Version]
- Posada, A.C.; Kolar, S.L.; Dusi, R.G.; Francois, P.; Roberts, A.A.; Hamilton, C.J.; Liu, G.Y.; Cheung, A. Importance of Bacillithiol in the Oxidative Stress Response of Staphylococcus aureus. Infect. Immun. 2014, 82, 316–332. [Google Scholar] [CrossRef] [Green Version]
- Fahey, R.C.; Brown, W.C.; Adams, W.B.; Worsham, M.B. Occurrence of glutathione in bacteria. J. Bacteriol. 1978, 133, 1126–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gout, I. Coenzyme A: A protective thiol in bacterial antioxidant defence. Biochem. Soc. Trans. 2019, 47, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, Y.; Peak-Chew, S.Y.; Newell, C.; Miller-Aidoo, S.; Mangal, S.; Zhyvoloup, A.; Bakovic’, J.; Malanchuk, O.; Pereira, G.C.; Kotiadis, V.; et al. Protein CoAlation: A redox-regulated protein modification by coenzyme A in mammalian cells. Biochem. J. 2017, 474, 2489–2508. [Google Scholar] [CrossRef] [Green Version]
- Tsuchiya, Y.; Zhyvoloup, A.; Baković, J.; Thomas, N.; Yu, B.Y.K.; Das, S.; Orengo, C.; Newell, C.; Ward, J.; Saladino, G.; et al. Protein CoAlation and antioxidant function of coenzyme A in prokaryotic cells. Biochem. J. 2018, 475, 1909–1937. [Google Scholar] [CrossRef] [Green Version]
- Delcardayré, S.B.; Stock, K.P.; Newton, G.L.; Fahey, R.C.; Davies, J.E. Coenzyme A Disulfide Reductase, the Primary Low Molecular Weight Disulfide Reductase from Staphylococcus aureus: Purification and characterization of the native enzyme. J. Biol. Chem. 1998, 273, 5744–5751. [Google Scholar] [CrossRef] [Green Version]
- Delcardayré, S.B.; Davies, J.E. Staphylococcus aureus Coenzyme A Disulfide Reductase, a New Subfamily of Pyridine Nucleotide-Disulfide Oxidoreductase Sequence, Expression, and Analysis of cdr. J. Biol. Chem. 1998, 273, 5752–5757. [Google Scholar] [CrossRef] [Green Version]
- Luba, J.; Charrier, V.; Claiborne, A. Coenzyme A-Disulfide Reductase from Staphylococcus aureus: Evidence for Asymmetric Behavior on Interaction with Pyridine Nucleotides. Biochemistry 1999, 38, 2725–2737. [Google Scholar] [CrossRef] [PubMed]
- Mikheyeva, I.V.; Thomas, J.M.; Kolar, S.L.; Corvaglia, A.; Gaϊa, N.; Leo, S.; Francois, P.; Liu, G.Y.; Rawat, M.; Cheung, A.L. YpdA, a putative bacillithiol disulfide reductase, contributes to cellular redox homeostasis and virulence in Staphylococcus aureus. Mol. Microbiol. 2019, 111, 1039–1056. [Google Scholar] [CrossRef] [PubMed]
- Linzner, N.; Van Loi, V.; Fritsch, V.N.; Tung, Q.N.; Stenzel, S.; Wirtz, M.; Hell, R.; Hamilton, C.J.; Tedin, K.; Fulde, M.; et al. Staphylococcus aureus Uses the Bacilliredoxin (BrxAB)/Bacillithiol Disulfide Reductase (YpdA) Redox Pathway to Defend Against Oxidative Stress Under Infections. Front. Microbiol. 2019, 10, 1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammerstad, M.; Gudim, I.; Hersleth, H.-P. The Crystal Structures of Bacillithiol Disulfide Reductase Bdr (YpdA) Provide Structural and Functional Insight into a New Type of FAD-Containing NADPH-Dependent Oxidoreductase. Biochemistry 2020, 59, 4793–4798. [Google Scholar] [CrossRef] [PubMed]
- Mallett, T.C.; Wallen, J.R.; Karplus, P.A.; Sakai, H.; Tsukihara, T.; Claiborne, A. Structure of Coenzyme A−Disulfide Reductase from Staphylococcus aureus at 1.54 Å Resolution, Biochemistry 2006, 45, 11278–11289. [Google Scholar] [CrossRef] [Green Version]
- Kowtoniuk, W.E.; Shen, Y.; Heemstra, J.M.; Agarwal, I.; Liu, D.R. A chemical screen for biological small molecule–RNA conjugates reveals CoA-linked RNA. Proc. Natl. Acad. Sci. USA 2009, 106, 7768–7773. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.G.; Kowtoniuk, W.E.; Agarwal, I.; Shen, Y.; Liu, D.R. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 2009, 5, 879–881. [Google Scholar] [CrossRef] [Green Version]
- Huang, F. Efficient incorporation of CoA, NAD and FAD into RNA by in vitro transcription. Nucleic Acids Res. 2003, 31, e8. [Google Scholar] [CrossRef] [Green Version]
- Bird, J.G.; Zhang, Y.; Tian, Y.; Panova, N.; Barvík, I.; Greene, L.; Liu, M.; Buckley, B.; Krasny, L.; Lee, J.K.; et al. The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 2016, 535, 444–447. [Google Scholar] [CrossRef] [Green Version]
- Julius, C.; Salgado, P.S.; Yuzenkova, Y. Metabolic cofactors NADH and FAD act as non-canonical initiating substrates for a primase and affect replication primer processing in vitro. Nucleic Acids Res. 2020, 48, 7298–7306. [Google Scholar] [CrossRef]
- Bird, J.G.; Basu, U.; Kuster, D.; Ramachandran, A.; Grudzien-Nogalska, E.; Towheed, A.; Wallace, D.C.; Kiledjian, M.; Temiakov, D.; Patel, S.S.; et al. Highly efficient 5’ capping of mitochondrial RNA with NAD+ and NADH by yeast and human mitochondrial RNA polymerase. eLife 2018, 7, e42179. [Google Scholar] [CrossRef] [PubMed]
- Höfer, K.; Li, S.; Abele, F.; Frindert, J.; Schlotthauer, J.; Grawenhoff, J.; Du, J.; Patel, D.J.; Jäschke, A. Structure and function of the bacterial decapping enzyme NudC. Nat. Chem. Biol. 2016, 12, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Li, K.; Huang, W.; Zhong, H.; Wu, H.; Wang, Y.; Zhang, H.; Cai, Z.; Guo, H.; Chen, X.; et al. Arabidopsis DXO1 possesses deNADding and exonuclease activities and its mutation affects defense-related and photosynthetic gene expression. J. Integr. Plant Biol. 2019, 62, 967–983. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Doamekpor, S.K.; Bird, J.G.; Nickels, B.E.; Tong, L.; Hart, R.P.; Kiledjian, M. 5′ End Nicotinamide Adenine Dinucleotide Cap in Human Cells Promotes RNA Decay through DXO-Mediated deNADding. Cell 2017, 168, 1015–1027.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Kuster, D.; Schmidt, T.; Kirrmaier, D.; Nübel, G.; Ibberson, D.; Benes, V.; Hombauer, H.; Knop, M.; Jäschke, A. Extensive 5′-surveillance guards against non-canonical NAD-caps of nuclear mRNAs in yeast. Nat. Commun. 2020, 11, 5508. [Google Scholar] [CrossRef]
- Doamekpor, S.K.; Grudzien-Nogalska, E.; Mlynarska-Cieslak, A.; Kowalska, J.; Kiledjian, M.; Tong, L. DXO/Rai1 enzymes remove 5′-end FAD and dephospho-CoA caps on RNAs. Nucleic Acids Res. 2020, 48, 6136–6148. [Google Scholar] [CrossRef]
- Sharma, S.; Grudzien-Nogalska, E.; Hamilton, K.; Jiao, X.; Yang, J.; Tong, L.; Kiledjian, M. Mammalian Nudix proteins cleave nucleotide metabolite caps on RNAs. Nucleic Acids Res. 2020, 48, 6788–6798. [Google Scholar] [CrossRef]
- Wang, J.; Chew, B.L.A.; Lai, Y.; Dong, H.; Xu, L.; Balamkundu, S.; Cai, W.M.; Cui, L.; Liu, C.F.; Fu, X.-Y.; et al. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 2019, 47, e130. [Google Scholar] [CrossRef] [Green Version]
- Grudzien-Nogalska, E.; Bird, J.G.; Nickels, B.E.; Kiledjian, M. “NAD-capQ” detection and quantitation of NAD caps. RNA 2018, 24, 1418–1425. [Google Scholar] [CrossRef] [Green Version]
- Walters, R.W.; Matheny, T.; Mizoue, L.S.; Rao, B.S.; Muhlrad, D.; Parker, R. Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2016, 114, 480–485. [Google Scholar] [CrossRef] [Green Version]
- Frindert, J.; Zhang, Y.; Nübel, G.; Kahloon, M.; Kolmar, L.; Hotz-Wagenblatt, A.; Burhenne, J.; Haefeli, W.E.; Jäschke, A. Identification, Biosynthesis, and Decapping of NAD-Capped RNAs in B. subtilis. Cell Rep. 2018, 24, 1890–1901.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, S.; Zhao, Y.; You, C.; Le, B.; Gong, Z.; Mo, B.; Xia, Y.; Chen, X. NAD+-capped RNAs are widespread in the Arabidopsis transcriptome and can probably be translated. Proc. Natl. Acad. Sci. USA 2019, 116, 12094–12102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales-Filloy, H.G.; Zhang, Y.; Nübel, G.; George, S.E.; Korn, N.; Wolz, C.; Jäschke, A. The 5′ NAD Cap of RNAIII Modulates Toxin Production in Staphylococcus aureus Isolates. J. Bacteriol. 2020, 202, e00591. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Flynn, N.; Chen, X. Discovery, Processing, and Potential Role of Noncanonical Caps in RNA. In Epitranscriptomics. RNA Technologies; Jurga, S., Barciszewski, J., Eds.; Springer: Cham, Switzerland, 2021; pp. 435–469. [Google Scholar] [CrossRef]
- Goosen, R.; Strauss, E. Simultaneous quantification of coenzyme A and its salvage pathway intermediates in in vitro and whole cell-sourced samples. RSC Adv. 2017, 7, 19717–19724. [Google Scholar] [CrossRef] [Green Version]
- Nübel, G.; Sorgenfrei, F.A.; Jäschke, A. Boronate affinity electrophoresis for the purification and analysis of cofactor-modified RNAs. Methods 2016, 117, 14–20. [Google Scholar] [CrossRef]
- Luciano, D.J.; Belasco, J.G. Analysis of RNA 5′ ends: Phosphate enumeration and cap characterization. Methods 2018, 155, 3–9. [Google Scholar] [CrossRef]
- Igloi, G.L. Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry 1988, 27, 3842–3849. [Google Scholar] [CrossRef]
- Spangler, J.R.; Huang, F. The E. coli NudL enzyme is a Nudix hydrolase that cleaves CoA and its derivatives. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhou, W.; Guan, Z.; Zhao, F.; Ye, Y.; Yang, F.; Yin, P.; Zhang, D. Structural insights into dpCoA-RNA decapping by NudC. RNA Biol. 2021, 18, 244–253. [Google Scholar] [CrossRef]
- Geerlof, A.; Lewendon, A.; Shaw, W.V. Purification and Characterization of Phosphopantetheine Adenylyltransferase from Escherichia coli. J. Biol. Chem. 1999, 274, 27105–27111. [Google Scholar] [CrossRef] [Green Version]
- Tsuchiya, Y.; Pham, U.; Gout, I. Methods for measuring CoA and CoA derivatives in biological samples. Biochem. Soc. Trans. 2014, 42, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.; Arias, N.; Acevedo, A.; Reddy, S.; Divakaruni, A.; Meriwether, D. A Single LC-MS/MS Analysis to Quantify CoA Biosynthetic Intermediates and Short-Chain Acyl CoAs. Metabolites 2021, 11, 468. [Google Scholar] [CrossRef] [PubMed]
- Duffy, E.E.; Rutenberg-Schoenberg, M.; Stark, C.D.; Kitchen, R.R.; Gerstein, M.; Simon, M.D. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Mol. Cell 2015, 59, 858–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhyvoloup, A.; Yu, B.Y.K.; Baković, J.; Davis-Lunn, M.; Tossounian, M.-A.; Thomas, N.; Tsuchiya, Y.; Peak-Chew, S.Y.; Wigneshweraraj, S.; Filonenko, V.; et al. Analysis of disulphide bond linkage between CoA and protein cysteine thiols during sporulation and in spores of Bacillus species. FEMS Microbiol. Lett. 2020, 367, fnaa174. [Google Scholar] [CrossRef] [PubMed]
- Sengle, G.; Jenne, A.; Arora, P.S.; Seelig, B.; Nowick, J.S.; Jäschke, A.; Famulok, M. Synthesis, incorporation efficiency, and stability of disulfide bridged functional groups at RNA 5′-ends. Bioorg. Med. Chem. 2000, 8, 1317–1329. [Google Scholar] [CrossRef]
- Wu, C.-W.; Eder, P.S.; Gopalan, V.; Behrman, E.J. Kinetics of Coupling Reactions That Generate Monothiophosphate Disulfides: Implications for Modification of RNAs. Bioconjug. Chem. 2001, 12, 842–844. [Google Scholar] [CrossRef] [PubMed]
- De Talancé, V.L.; Bauer, F.; Hermand, D.; Vincent, S. A simple synthesis of APM ([p-(N-acrylamino)-phenyl] mercuric chloride), a useful tool for the analysis of thiolated biomolecules. Bioorg. Med. Chem. Lett. 2011, 21, 7265–7267. [Google Scholar] [CrossRef]
- Igloi, G.L.; Kössel, H. Affinity electrophoresis for monitoring terminal phosphorylation and the presence of queuosine in RNA. Application of polyacrylamide containing a covalently bound boronic acid. Nucleic Acids Res. 1985, 13, 6881–6898. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, A.O.; Jacobsen, J. Reactivity of the Thiol Group in Human and Bovine Albumin at pH 3-9, as Measured by Exchange with 2,2′-Dithiodipyridine. JBIC J. Biol. Inorg. Chem. 1980, 106, 291–295. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Löcherer, C.; Bühler, N.; Lafrenz, P.; Jäschke, A. Staphylococcus aureus Small RNAs Possess Dephospho-CoA 5′-Caps, but No CoAlation Marks. Non-Coding RNA 2022, 8, 46. https://doi.org/10.3390/ncrna8040046
Löcherer C, Bühler N, Lafrenz P, Jäschke A. Staphylococcus aureus Small RNAs Possess Dephospho-CoA 5′-Caps, but No CoAlation Marks. Non-Coding RNA. 2022; 8(4):46. https://doi.org/10.3390/ncrna8040046
Chicago/Turabian StyleLöcherer, Christian, Nadja Bühler, Pascal Lafrenz, and Andres Jäschke. 2022. "Staphylococcus aureus Small RNAs Possess Dephospho-CoA 5′-Caps, but No CoAlation Marks" Non-Coding RNA 8, no. 4: 46. https://doi.org/10.3390/ncrna8040046