Quantitative Analysis of Genomic DNA Degradation of E. coli Using Automated Gel Electrophoresis under Various Levels of Microwave Exposure
Abstract
:1. Introduction
2. Results and Discussion
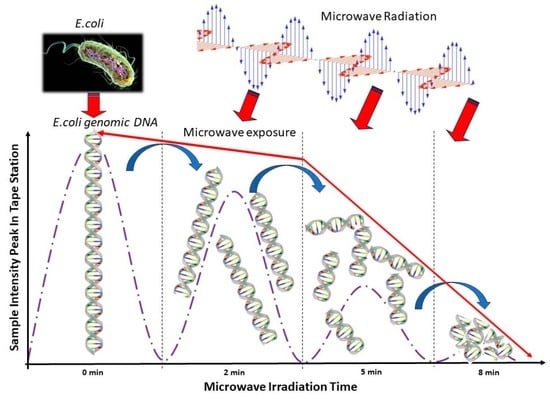
2.1. Automated Electrophoresis and Electropherograms Using Automated Electrophoresis under Various Microwave Exposure Conditions
2.2. E. coli Enumeration and Corresponding Automated Electropherogram Analysis
2.3. Comparison between Automated Electrophoresis and Qubit Fluorometer Measurements
3. Conclusions
4. Material and Methods
4.1. E. coli Isolation and Membrane Filtration Method
4.2. Genomic DNA Extraction from Fecal Thermotolerant E. coli
4.3. Automated Genomic DNA Integrity Evaluation of E. coli
4.4. Microwave Experiment for E. coli DNA Degradation and Genomic DNA Monitoring
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, P.Y.; Costumbrado, J.; Hsu, C.Y.; Kim, Y.H. Agarose gel electrophoresis for the separation of DNA fragments. JoVE 2012, 62, e3923. [Google Scholar]
- Sowersby, D.S.; Lewis, L.K. SURE gel electrophoresis: A method for improved detection and purification of dilute nucleic acid samples. Anal. Biochem. 2024, 684, 115373. [Google Scholar] [CrossRef] [PubMed]
- Terabayashi, T.; Tokumaru, A.; Ishizaki, T.; Hanada, K. Analysis of chromosomal DNA fragmentation in apoptosis by pulsed-field gel electrophoresis. In DNA Electrophoresis: Methods and Protocols; Humana: New York, NY, USA, 2020; pp. 89–99. [Google Scholar]
- Wang, Z.; Wang, P.; Tao, C.; Zhang, D.; Li, Z.; Yamaguchi, Y. Capillary electrophoresis of DNA with high resolution based on copoly (pentaerythritoltetra succinimidylcarboxypentyl/aminopropyl polyoxyethylene) hydrogel. Anal. Chim. Acta 2021, 1178, 338811. [Google Scholar] [CrossRef] [PubMed]
- Dangerfield, T.L.; Huang, N.Z.; Johnson, K.A. High throughput quantification of short nucleic acid samples by capillary electrophoresis with automated data processing. Anal. Biochem. 2021, 629, 114239. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Polyacrylamide gel electrophoresis. Cold Spring Harb. Protoc. 2020, 2020, pdb-rot100412. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.B.; Aldridge, P.K.; Callis, J.B. Observation of individual DNA molecules undergoing gel electrophoresis. Science 1989, 243, 203–206. [Google Scholar] [CrossRef]
- Aaij, C.; Borst, P. The gel electrophoresis of DNA. Biochim. Et Biophys. Acta (BBA)-Nucleic Acids Protein Synth. 1972, 269, 192–200. [Google Scholar] [CrossRef]
- Helling, R.B.; Goodman, H.M.; Boyer, H.W. Analysis of endonuclease R–EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J. Virol. 1974, 14, 1235–1244. [Google Scholar] [CrossRef] [PubMed]
- Elliott, C.I.; Stotesbury, T.E.; Shafer, A.B. Using total RNA quality metrics for time since deposition estimates in degrading bloodstains. J. Forensic Sci. 2022, 67, 1776–1785. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, Y.; Yamaguchi, H.; Einaga, N.; Esumi, M. Pitfalls of DNA quantification using DNA-binding fluorescent dyes and suggested solutions. PLoS ONE 2016, 11, e0150528. [Google Scholar] [CrossRef] [PubMed]
- Masago, K.; Fujita, S.; Oya, Y.; Takahashi, Y.; Matsushita, H.; Sasaki, E.; Kuroda, H. Comparison between fluorimetry (Qubit) and spectrophotometry (NanoDrop) in the quantification of DNA and RNA extracted from frozen and FFPE tissues from lung cancer patients: A real-world use of genomic tests. Medicina 2021, 57, 1375. [Google Scholar] [CrossRef]
- Robin, J.D.; Ludlow, A.T.; LaRanger, R.; Wright, W.E.; Shay, J.W. Comparison of DNA quantification methods for next generation sequencing. Sci. Rep. 2016, 6, 24067. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. Comparison of methods for the quantification of cell-free DNA isolated from cell culture supernatant. Tumor Biol. 2019, 41, 1010428319866369. [Google Scholar] [CrossRef]
- Oswald, J.A.; Allen, J.M.; LeFebvre, M.J.; Stucky, B.J.; Folk, R.A.; Albury, N.A.; Morgan, G.S.; Guralnick, R.P.; Steadman, D.W. Ancient DNA and high-resolution chronometry reveal a long-term human role in the historical diversity and biogeography of the Bahamian hutia. Sci. Rep. 2020, 10, 1373. [Google Scholar] [CrossRef]
- Brena, R.M.; Auer, H.; Kornacker, K.; Hackanson, B.; Raval, A.; Byrd, J.C.; Plass, C. Accurate quantification of DNA methylation using combined bisulfite restriction analysis coupled with the Agilent 2100 Bioanalyzer platform. Nucleic Acids Res. 2006, 34, e17. [Google Scholar] [CrossRef]
- Panaro, N.J.; Yuen, P.K.; Sakazume, T.; Fortina, P.; Kricka, L.J.; Wilding, P. Evaluation of DNA fragment sizing and quantification by the agilent 2100 bioanalyzer. Clin. Chem. 2000, 46, 1851–1853. [Google Scholar] [CrossRef] [PubMed]
- Vitale, D. Comparing the Agilent 2100 Bioanalyzer Performance to Traditional DNA Analysis Techniques; Agilent Technologies Publication: Santa Clara, CA, USA, 2000. [Google Scholar]
- Naphade, S.; Bhatnagar, R.; Hanson-Smith, V.; Choi, I.; Zhang, A. Systematic comparative analysis of strand-specific RNA-seq library preparation methods for low input samples. Sci. Rep. 2022, 12, 1789. [Google Scholar] [CrossRef] [PubMed]
- Trinidad, E.M.; Juan-Ribelles, A.; Pisano, G.; Castel, V.; Cañete, A.; Gut, M.; Heath, S.; Font de Mora, J. Evaluation of circulating tumor DNA by electropherogram analysis and methylome profiling in high-risk neuroblastomas. Front. Oncol. 2023, 13, 1037342. [Google Scholar] [CrossRef] [PubMed]
- Mudhoo, A.; Sharma, S.K. Microwave irradiation technology in waste sludge and wastewater treatment research. Crit. Rev. Environ. Sci. Technol. 2011, 41, 999–1066. [Google Scholar] [CrossRef]
- Remya, N.; Lin, J.G. Current status of microwave application in wastewater treatment—A review. Chem. Eng. J. 2011, 166, 797–813. [Google Scholar] [CrossRef]
- Wang, C.; Song, L.; Zhang, Z.; Wang, Y.; Xie, X. Microwave-induced release and degradation of airborne antibiotic resistance genes (ARGs) from Escherichia coli bioaerosol based on microwave absorbing material. J. Hazard. Mater. 2020, 394, 122535. [Google Scholar] [CrossRef] [PubMed]
- Kipcak, A.S.; Doymaz, I. Mathematical modeling and drying characteristics investigation of black mulberry dried by microwave method. Int. J. Fruit Sci. 2020, 20, S1222–S1233. [Google Scholar] [CrossRef]
- Lewicki, P.P. Design of hot air drying for better foods. Trends Food Sci. Technol. 2006, 17, 153–163. [Google Scholar] [CrossRef]
- Schiffmann, R.F. Microwave and dielectric drying. In Handbook of Industrial Drying; CRC Press: Boca Raton, FL, USA, 2020; pp. 345–372. [Google Scholar]
- Zhang, Y.; Li, M.; Liu, Y.; Guan, E.; Bian, K. Degradation of aflatoxin B1 by water-assisted microwave irradiation: Kinetics, products, and pathways. LWT 2021, 152, 112310. [Google Scholar] [CrossRef]
- Davis, M.E.; VanZandt, L.L. Microwave response of DNA in solution: Theory. Phys. Rev. A 1988, 37, 888. [Google Scholar] [CrossRef] [PubMed]
- Banik, S.B.; Bandyopadhyay, S.; Ganguly, S. Bioeffects of microwave—A brief review. Bioresour. Technol. 2003, 87, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Alkis, M.E.; Akdag, M.Z.; Dasdag, S. Effects of low-intensity Microwave Radiation on Oxidant-antioxidant Parameters and DNA Damage in the Liver of rats. Bioelectromagnetics 2021, 42, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Pajares, M.J.; Palanca-Ballester, C.; Urtasun, R.; Alemany-Cosme, E.; Lahoz, A.; Sandoval, J. Methods for analysis of specific DNA methylation status. Methods 2021, 187, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Tozzo, P.; Scrivano, S.; Sanavio, M.; Caenazzo, L. The role of DNA degradation in the estimation of post-mortem interval: A systematic review of the current literature. Int. J. Mol. Sci. 2020, 21, 3540. [Google Scholar] [CrossRef] [PubMed]
- Abdulla, J.M.; Gomaa, R.; Attalla, S.M.; Nader, L.M. Investigation of DNA degradation in forensic blood samples after exposure to different environmental conditions. Int. J. Med. Toxicol. Leg. Med. 2021, 24, 66–74. [Google Scholar] [CrossRef]
- Lozano-Peral, D.; Rubio, L.; Santos, I.; Gaitán, M.J.; Viguera, E.; Martín-de-Las-Heras, S. DNA degradation in human teeth exposed to thermal stress. Sci. Rep. 2021, 11, 12118. [Google Scholar] [CrossRef] [PubMed]
- Jo, T.; Tsuri, K.; Hirohara, T.; Yamanaka, H. Warm temperature and alkaline conditions accelerate environmental RNA degradation. Environ. DNA 2022, 5, 836–848. [Google Scholar] [CrossRef]
- Saito, T.; Doi, H. Effect of salinity and water dilution on environmental DNA degradation in freshwater environments. bioRXiv 2021, 2021-05. [Google Scholar]
- Sato, S.; Takesono, S.; Imada, G. Inactivation of bacteria in water by pulsed power injection. IEEJ Trans. Electr. Electron. Eng. 2020, 15, 1400–1402. [Google Scholar] [CrossRef]
- Wang, X.; Son, A. Effects of pretreatment on the denaturation and fragmentation of genomic DNA for DNA hybridization. Environ. Sci. Process. Impacts 2013, 15, 2204–2212. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Lv, X.; Meng, L.; Ai, M.; Li, C.; Teng, F.; Feng, Z. Effect of microwave sterilization on maturation time and quality of low-salt sufu. Food Sci. Nutr. 2020, 8, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Hock, T.K.; Chala, G.T.; Cheng, H.H. An innovative hybrid steam-microwave sterilization of palm oil fruits at atmospheric pressure. Innov. Food Sci. Emerg. Technol. 2020, 60, 102289. [Google Scholar] [CrossRef]
- Swicord, M.L.; Davis, C.C. Microwave absorption of DNA between 8 and 12 GHz. Biopolym. Orig. Res. Biomol. 1982, 21, 2453–2460. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.; Kumar, N.; Mumtaz, S.; Lim, J.S.; Jang, J.H.; Kim, D.; Sahu, B.D.; Bogaerts, A.; Choi, E.H. Evaluation of non-thermal effect of microwave radiation and its mode of action in bacterial cell inactivation. Sci. Rep. 2021, 11, 14003. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Parhi, A.; Al-Ghamdi, S.; Sonar, C.R.; Mattinson, D.S.; Tang, J.; Yang, T.; Sablani, S.S. Stability of vitamin C, color, and garlic aroma of garlic mashed potatoes in polymer packages processed with microwave-assisted thermal sterilization technology. J. Food Sci. 2020, 85, 2843–2851. [Google Scholar] [CrossRef] [PubMed]
- Soni, A.; Smith, J.; Thompson, A.; Brightwell, G. Microwave-induced thermal sterilization-A review on history, technical progress, advantages and challenges as compared to the conventional methods. Trends Food Sci. Technol. 2020, 97, 433–442. [Google Scholar] [CrossRef]
- Heiman, K.E.; Mody, R.K.; Johnson, S.D.; Griffin, P.M.; Gould, L.H. Escherichia coli O157 outbreaks in the United States, 2003–2012. Emerg. Infect. Dis. 2015, 21, 1293–1301. [Google Scholar] [CrossRef]
- Rangel, J.M.; Sparling, P.H.; Crowe, C.; Griffin, P.M.; Swerdlow, D.L. Epidemiology of Escherichia coli O157: H7 outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 2005, 11, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Marshall, K.E.; Hexemer, A.; Seelman, S.L.; Fatica, M.K.; Blessington, T.; Hajmeer, M.; Kisselburgh, H.; Atkinson, R.; Hill, K.; Sharma, D.; et al. Lessons learned from a decade of investigations of Shiga toxin–producing Escherichia coli outbreaks linked to leafy greens, United States and Canada. Emerg. Infect. Dis. 2020, 26, 2319–2328. [Google Scholar] [CrossRef] [PubMed]
- Rahi, G.S.; Lodhi, K.; Grier, R.; Adams, J.; Torrez, A. Effects of microwave radiation on human DNA degradation. World Res. J. Appl. Phys. 2012, 3, 36–40. [Google Scholar]
- Wang, J.J.; Zhou, Y.Y.; Xiang, J.L.; Du, H.S.; Zhang, J.; Zheng, T.G.; Liu, M.; Ye, M.Q.; Chen, Z.; Du, Y. Disinfection of wastewater by a complete equipment based on a novel ultraviolet light source of microwave discharge electrodeless lamp: Characteristics of bacteria inactivation, reactivation and full-scale studies. Sci. Total Environ. 2024, 917, 170200. [Google Scholar] [CrossRef]
- Mori, I.; Ozaki, T.; Tabuse, K.; Utsunomiya, H.; Taniguchi, E.; Kakudo, K. Microwave cell death: Molecular analysis using DNA electrophoresis, PCR amplification and TUNEL. Pathol. Int. 2009, 59, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Pratiwi, N.; Pramila, C.; Safitri, F.; Namidya, S.K.; Putri, D.H. The Effect of Micro Radiation on Microbial DNA. J. Serambi Biol. 2023, 8, 74–78. [Google Scholar]
- Prudhvi, P.V.; Kar, S.; Sharma, P.; Patel, J.; Nayak, S.P. Synergistic effect of sonication and microwave for inhibition of microorganism. In Ultrasound and Microwave for Food Processing; Academic Press: Cambridge, MA, USA, 2023; pp. 189–216. [Google Scholar]
- Wilson-Garner, S.; Alzeer, S.; Baillie, L.; Porch, A. High-volume biological sample processing using microwaves. J. Appl. Phys. 2024, 135, 044901. [Google Scholar] [CrossRef]
- Sharma, P.; Venugopal, A.P.; Sutar, P.P.; Xiao, H.; Zhang, Q. Mechanism of microbial spore inactivation through electromagnetic radiations: A review. J. Future Foods 2024, 4, 324–334. [Google Scholar] [CrossRef]
- Zlotorzynski, A. The application of microwave radiation to analytical and environmental chemistry. Crit. Rev. Anal. Chem. 1995, 25, 43–76. [Google Scholar] [CrossRef]
- Kostas, E.T.; Beneroso, D.; Robinson, J.P. The application of microwave heating in bioenergy: A review on the microwave pre-treatment and upgrading technologies for biomass. Renew. Sustain. Energy Rev. 2017, 77, 12–27. [Google Scholar] [CrossRef]
- Santagada, V.; Perissutti, E.; Caliendo, G. The application of microwave irradiation as new convenient synthetic procedure in drug discovery. Curr. Med. Chem. 2002, 9, 1251–1283. [Google Scholar] [CrossRef] [PubMed]
- Lovás, M.; Znamenáčková, I.; Zubrik, A.; Kováčová, M.; Dolinská, S. The application of microwave energy in mineral processing—A review. Acta Montan. Slovaca 2011, 16, 137–148. [Google Scholar]
- Proestos, C.; Komaitis, M. Application of microwave-assisted extraction to the fast extraction of plant phenolic compounds. LWT-Food Sci. Technol. 2008, 41, 652–659. [Google Scholar] [CrossRef]
- Whittaker, A.G.; Mingos, D.M. The application of microwave heating to chemical syntheses. J. Microw. Power Electromagn. Energy 1994, 29, 195–219. [Google Scholar] [CrossRef]
- Baghel, P.K. Application of microwave in manufacturing technology: A review. Mater. Today Proc. 2023, in press. [Google Scholar]
- Zhang, L.; Zhang, Z.; Yu, W.; Miao, Y. Review of the Application of Microwave Heating Technology in Asphalt Pavement Self-Healing and De-icing. Polymers 2023, 15, 1696. [Google Scholar] [CrossRef] [PubMed]
- Strekalova, A.A.; Shesterkina, A.A.; Kustov, A.L.; Kustov, L.M. Recent Studies on the Application of Microwave-Assisted Method for the Preparation of Heterogeneous Catalysts and Catalytic Hydrogenation Processes. Int. J. Mol. Sci. 2023, 24, 8272. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Lou, X.; Wang, Y.; Yang, S.; Li, Z.; Zhang, X.; Yan, Y.; Xin, H. A Review on the Application of Microwave Absorbents in Oil Shale. Ind. Eng. Chem. Res. 2023, 62, 19402–19426. [Google Scholar] [CrossRef]
- Luo, S.; Ge, Y.; Zhao, M.; Yang, L.; Ren, J. Application of microwave hydrothermal synthesis for the solidification of copper: Effect of heavy metal content and microwave time. Process Saf. Environ. Prot. 2023, 173, 765–774. [Google Scholar] [CrossRef]
- Kabir, E. Application of microwave heating in polymer synthesis: A review. Results Chem. 2023, 6, 101178. [Google Scholar] [CrossRef]
- EPA/821/R-97/004; Improved Enumeration Methods for the Recreational Water Quality Indicators: Enterococci and Escherichia coli. United States Environmental Protection Agency (US EPA): Washington, DC, USA, 2000.









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, A.; Momeni, O.; Pandey, P. Quantitative Analysis of Genomic DNA Degradation of E. coli Using Automated Gel Electrophoresis under Various Levels of Microwave Exposure. Gels 2024, 10, 242. https://doi.org/10.3390/gels10040242
Pandey A, Momeni O, Pandey P. Quantitative Analysis of Genomic DNA Degradation of E. coli Using Automated Gel Electrophoresis under Various Levels of Microwave Exposure. Gels. 2024; 10(4):242. https://doi.org/10.3390/gels10040242
Chicago/Turabian StylePandey, Aditya, Omeed Momeni, and Pramod Pandey. 2024. "Quantitative Analysis of Genomic DNA Degradation of E. coli Using Automated Gel Electrophoresis under Various Levels of Microwave Exposure" Gels 10, no. 4: 242. https://doi.org/10.3390/gels10040242