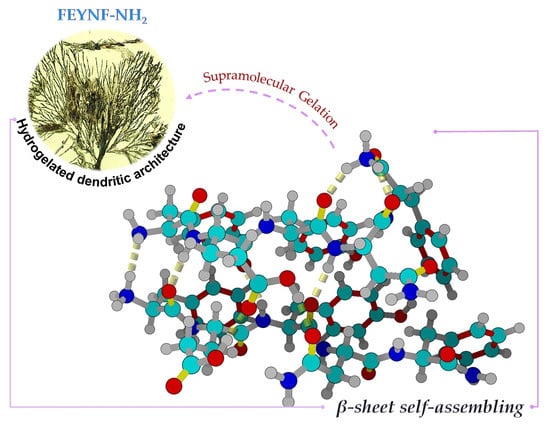
Self-Assembly of a Novel Pentapeptide into Hydrogelated Dendritic Architecture: Synthesis, Properties, Molecular Docking and Prospective Applications
Abstract
:1. Introduction
2. Results and Discussion
2.1. Solid-Phase Synthesis of Novel FEYNF-NH2 Pentapeptide
2.2. Qualitative FEYNF-NH2 Peptide Analysis and Purification
2.3. Molecular Mass Confirmation (MALDI-ToF MS and MS/MS)
2.4. Self-Aggregation of FEYNF-NH2 Investigations
2.4.1. Fourier-Transform Infrared Spectroscopy (FTIR)
2.4.2. Fluorescence Studies
2.4.3. Transmission Electron Microscopy (TEM)
2.4.4. Polarized Optical Light Microscopy (POM)
2.5. Molecular Docking Simulation
2.5.1. Insights into FEYNF-NH2 Self-Assembling Mechanism
2.5.2. Investigation of β-Sheet Structure Formation
2.6. Potential Use of FEYNF Self-Assembling Aggregates in Polyplex Formation
3. Conclusions
4. Materials and Methods
4.1. Reagents
4.2. Solid-Phase Synthesis of FEYNF-NH2 Pentapeptide
4.3. Reverse-Phase High-Performance Liquid Chromatography
4.4. Solid-Phase Extraction of FEYNF-NH2 Peptide
4.5. Mass Spectrometry Analysis (MALDI-ToF MS and MS/MS)
4.6. Fluorescence Analysis
4.7. Sample Preparation Protocol for Incubation in Physiological Conditions
4.8. Molecular Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, B.; Tao, K.; Jia, Y.; Yan, X.; Zou, Q.; Gazit, E.; Li, J. Photoactive properties of supramolecular assembled short peptides. Chem. Soc. Rev. 2019, 48, 4387–4400. [Google Scholar] [CrossRef] [PubMed]
- Abbas, M.; Ovais, M.; Atiq, A.; Ansari, T.M.; Xing, R.; Spruijt, E.; Yan, X. Tailoring supramolecular short peptide nanomaterials for antibacterial applications. Coord. Chem. Rev. 2022, 460, 214481. [Google Scholar] [CrossRef]
- Ellert-Miklaszewska, A.; Poleszak, K.; Kaminska, B. Short peptides interfering with signaling pathways as new therapeutic tools for cancer treatment. Future Med. Chem. 2017, 9, 199–221. [Google Scholar] [CrossRef]
- Restu, W.K.; Yamamoto, S.; Nishida, Y.; Ienaga, H.; Aoi, T.; Maruyama, T. Hydrogel formation by short D-peptide for cell-culture scaffolds. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 111, 110746. [Google Scholar] [CrossRef]
- Ross, A.; Sauce-Guevara, M.A.; Alarcon, E.I.; Mendez-Rojas, M.A. Peptide Biomaterials for Tissue Regeneration. Front. Bioeng. Biotechnol. 2022, 10, 893936. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, M.; Wang, T.; Sun, M.; Huang, H.; Shi, X.; Duan, S.; Wu, Y.; Zhu, J.; Liu, F. Self-assembled short peptides: Recent advances and strategies for potential pharmaceutical applications. Mater. Today Bio 2023, 20, 100644. [Google Scholar] [CrossRef] [PubMed]
- Sfragano, P.S.; Moro, G.; Polo, F.; Palchetti, I. The Role of Peptides in the Design of Electrochemical Biosensors for Clinical Diagnostics. Biosensors 2021, 11, 246. [Google Scholar] [CrossRef]
- Riley, M.K.; Vermerris, W. Recent Advances in Nanomaterials for Gene Delivery—A Review. Nanomaterials 2017, 7, 94. [Google Scholar] [CrossRef]
- Tarvirdipour, S.; Huang, X.; Mihali, V.; Schoenenberger, C.A.; Palivan, C.G. Peptide-Based Nanoassemblies in Gene Therapy and Diagnosis: Paving the Way for Clinical Application. Molecules 2020, 25, 3482. [Google Scholar] [CrossRef]
- Habibi, N.; Kamaly, N.; Memic, A.; Shafiee, H. Self-assembled peptide-based nanostructures: Smart nanomaterials toward targeted drug delivery. Nano Today 2016, 11, 41–60. [Google Scholar] [CrossRef]
- Wang, W.; Hu, Z. Targeting Peptide-Based Probes for Molecular Imaging and Diagnosis. Adv. Mater. 2019, 31, 1804827. [Google Scholar] [CrossRef] [PubMed]
- Petit, N.; Dyer, J.M.; Gerrard, J.A.; Domigan, L.J.; Clerens, S. Insight into the self-assembly and gel formation of a bioactive peptide derived from bovine casein. BBA Adv. 2023, 3, 100086. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xie, L.; Zheng, R.; Sun, R. Self-Assembly Dipeptide Hydrogel: The Structures and Properties. Front. Chem. 2021, 9, 739791. [Google Scholar] [CrossRef] [PubMed]
- Webber, M.J.; Pashuck, E.T. (Macro)molecular Self-Assembly for Hydrogel Drug Delivery. Adv. Drug Deliv. Rev. 2021, 172, 275–295. [Google Scholar] [CrossRef]
- Hansen, P.R.; Oddo, A. Methods in Molecular Biology; Fmoc Solid-Phase Peptide Synthesis. In Peptide Antibodies; Houen, G., Ed.; Humana Press: New York, NY, USA, 2015; Volume 1348, pp. 33–50. [Google Scholar] [CrossRef]
- Behrendt, R.; White, P.; Offer, J. Advances in Fmoc solid-phase peptide synthesis. J. Pept. Sci. 2016, 22, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Tang, Y.; Yamamoto, T.; Yao, Y.; Guterman, T.; Zilberzwige-Tal, S.; Adadi, N.; Ji, W.; Dvir, T.; Ramamoorthy, A.; et al. Unusual Two-Step Assembly of a Minimalistic Dipeptide-Based Functional Hypergelator. Adv. Mater. 2020, 32, 1906043. [Google Scholar] [CrossRef] [PubMed]
- Seow, W.Y.; Hauser, C.A. Short to ultrashort peptide hydrogels for biomedical uses. Mater. Today 2014, 17, 381–388. [Google Scholar] [CrossRef]
- Martin, V.; Egelund, P.H.G.; Johansson, H.; Le Quement, S.T.; Wojcik, F.; Pedersen, D.S. Greening the synthesis of peptide therapeutics: An industrial perspective. RSC Adv. 2020, 10, 42457–42492. [Google Scholar] [CrossRef]
- Gazit, E. Self-Assembled Peptide Nanostructures: The Design of Molecular Building Blocks and Their Technological Utilization. Chem. Soc. Rev. 2007, 36, 1263–1269. [Google Scholar] [CrossRef]
- Handelman, A. Optical Polarization-Based Measurement Methods for Characterization of Self-Assembled Peptides’ and Amino Acids’ Micro- and Nanostructures. Molecules 2022, 27, 1802. [Google Scholar] [CrossRef]
- Zhou, P.; Yuan, C.; Yan, X. Computational approaches for understanding and predicting the self-assembled peptide hydrogels. Curr. Opin. Colloid Interface Sci. 2022, 62, 101645. [Google Scholar] [CrossRef]
- Jitaru, S.-C.; Neamtu, A.; Drochioiu, G.; Darie-Ion, L.; Stoica, I.; Petre, B.-A.; Gradinaru, V.-R. A Diphenylalanine Based Pentapeptide with Fibrillating Self-Assembling Properties. Pharmaceutics 2023, 15, 371. [Google Scholar] [CrossRef] [PubMed]
- Lucas, X.; Bauzá, A.; Frontera, A.; Quiñonero, D. A thorough anion–π interaction study in biomolecules: On the importance of cooperativity effects. Chem. Sci. 2016, 7, 1038–1050. [Google Scholar] [CrossRef] [PubMed]
- Amblard, M.; Fehrentz, J.A.; Martinez, J.; Subra, G. Methods and protocols of modern solid phase peptide synthesis. Mol. Biotechnol. 2006, 33, 239–254. [Google Scholar] [CrossRef] [PubMed]
- Wellings, D.A.; Atherton, E. Standard Fmoc protocols. Methods Enzymol. 1997, 289, 44–67. [Google Scholar] [CrossRef] [PubMed]
- Arispe, N.; Diaz, J.C.; Flora, M. Efficiency of histidine-associating compounds for blocking the Alzheimer’s Aβ channel activity and cytotoxicity. Biophys. J. 2008, 95, 4879–4889. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, F.; Aróstica, M.; Román, T.; Beltrán, D.; Gauna, A.; Albericio, F.; Cárdenas, C. Peptides, solid-phase synthesis and characterization: Tailor-made methodologies. Electron. J. Biotechnol. 2023, 64, 27–33. [Google Scholar] [CrossRef]
- Insuasty Cepeda, D.S.; Pineda Castañeda, H.M.; Rodríguez Mayor, A.V.; García Castañeda, J.E.; Maldonado Villamil, M.; Fierro Medina, R.; Rivera Monroy, Z.J. Synthetic peptide purification via solid-phase extraction with gradient elution: A simple, economical, fast, and efficient methodology. Molecules 2019, 24, 1215. [Google Scholar] [CrossRef]
- Ruderman, G.; Caffarena, E.R.; Mogilner, I.G.; Tolosa, E.J. Hydrogen Bonding of carboxylic acids in aqueous solutions—UV spectroscopy, viscosity, and molecular simulation of acetic acid. J. Solut. Chem. 1998, 27, 935–948. [Google Scholar] [CrossRef]
- Körsgen, M.; Pelster, A.; Dreisewerd, K.; Arlinghaus, H.F. 3D ToF-SIMS analysis of peptide incorporation into MALDI matrix crystals with sub-micrometer resolution. J. Am. Soc. Mass Spectrom. 2015, 27, 277–284. [Google Scholar] [CrossRef]
- Moyer, S.C.; VonSeggern, C.E.; Cotter, R.J. Fragmentation of cationized phosphotyrosine containing peptides by atmospheric pressure MALDI/Ion trap mass spectrometry. J. Am. Soc. Mass Spectrom. 2003, 14, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Jitaru, S.C.; Enache, A.C.; Drochioiu, G.; Petre, B.A.; Gradinaru, V.R. Dendritic-like Self-Assembling Pentapeptide with Potential Applications in Emerging Biotechnologies. In Proceedings of the IFMBE Proceedings Series, Bucharest, Romania, 9–10 November 2023. [Google Scholar]
- Zhang, Z. Prediction of low-energy collision-induced dissociation spectra of peptides. Anal. Chem. 2004, 76, 3908–3922. [Google Scholar] [CrossRef] [PubMed]
- Bagińska, K.; Makowska, J.; Wiczk, W.; Kasprzykowski, F.; ChmurzyńSKI, L. Conformational studies of alanine-rich peptide using CD and FTIR spectroscopy. J. Pept. Sci. 2008, 14, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Hoffmann, W.; Warnke, S.; Huang, X.; Gewinner, S.; Schöllkopf, W.; Bowere, M.T.; von Helden, G.; Pagel, K. An infrared spectroscopy approach to follow β-sheet formation in peptide amyloid assemblies. Nat. Chem. 2016, 9, 39–44. [Google Scholar] [CrossRef] [PubMed]
- De Meutter, J.; Goormaghtigh, E. Amino acid side chain contribution to protein FTIR spectra: Impact on secondary structure evaluation. Eur. Biophys. J. 2021, 50, 641–651. [Google Scholar] [CrossRef]
- Barreto, M.S.C.; Elzinga, E.J.; Alleoni, L.R.F. The molecular insights into protein adsorption on hematite surface disclosed by in-situ ATR-FTIR/2D-COS study. Sci. Rep. 2020, 10, 13441. [Google Scholar] [CrossRef] [PubMed]
- Angelerou, M.G.F.; Markus, R.; Paraskevopoulou, V.; Foralosso, R.; Clarke, P.; Alvarez, C.V.; Chenlo, M.; Johnson, L.; Rutland, C.; Allen, S.; et al. Mechanistic investigations into the encapsulation and release of small molecules and proteins from a supramolecular nucleoside gel in vitro and in vivo. J. Control. Release 2020, 317, 118–129. [Google Scholar] [CrossRef]
- Creasey, R.C.; Mostert, A.B.; Solemanifar, A.; Nguyen, T.A.; Virdis, B.; Freguia, S.; Laycock, B. Biomimetic peptide nanowires designed for conductivity. ACS Omega 2019, 4, 1748–1756. [Google Scholar] [CrossRef]
- Radomska, K.; Wolszczak, M. Spontaneous and Ionizing Radiation-Induced Aggregation of Human Serum Albumin: Dityrosine as a Fluorescent Probe. Int. J. Mol. Sci. 2022, 23, 8090. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.; Mu, G.; Yang, L.; Ren, C.; Wang, Z.; Guo, Q.; Liu, J.; Yang, C. Structure of self-assembled peptide determines the activity of aggregation-induced emission luminogen-peptide conjugate for detecting alkaline phosphatase. Anal. Chem. 2022, 94, 2236–2243. [Google Scholar] [CrossRef]
- Xiong, Y.; Shi, C.; Li, L.; Tang, Y.; Zhang, X.; Liao, S.; Zhang, B.; Sun, C.; Ren, C. A review on recent advances in amino acid and peptide-based fluorescence and its potential applications. New J. Chem. 2021, 45, 15180–15194. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2013. [Google Scholar]
- Konda, M.; Bhowmik, S.; Mobin, S.M.; Biswas, S.; Das, A.K. Modulating Hydrogen Bonded Self-Assembled Patterns and Morphological Features by a Change in Side Chain of Third Amino Acid of Synthetic γ-Amino Acid Based Tripeptides. ChemistrySelect 2016, 1, 2586–2593. [Google Scholar] [CrossRef]
- Gelain, F.; Luo, Z.; Zhang, S. Self-Assembling Peptide EAK16 and RADA16 Nanofiber Scaffold Hydrogel. Chem. Rev. 2020, 120, 13434–13460. [Google Scholar] [CrossRef] [PubMed]
- DiCostanzo, A.C.; Thompson, J.R.; Peterson, F.C.; Volkman, B.F.; Ramirez-Alvarado, M. Tyrosine residues mediate fibril formation in a dynamic light chain dimer interface. JBC 2012, 287, 27997–28006. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Viles, J.H. pH Dependence of Amyloid-β Fibril Assembly Kinetics: Unravelling the Microscopic Molecular Processes. Angew. Chem. Int. Ed. 2022, 61, e202210675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Greenfield, M.A.; Mata, A.; Palmer, L.; Bitton, R.; Mantei, J.R.; Aparicio, C.; de la Cruz, M.O.; Stupp, S.I. A self-assembly pathway to aligned monodomain gels. Nat. Mater. 2010, 9, 594–601. [Google Scholar] [CrossRef]
- Lembré, P.; Di Martino, P.; Vendrely, C. Amyloid peptides derived from CsgA and FapC modify the viscoelastic properties of biofilm model matrices. Biofouling 2014, 30, 415–426. [Google Scholar] [CrossRef]
- Dapson, R.W. Amyloid from a histochemical perspective. A review of the structure, properties and types of amyloid, and a proposed staining mechanism for Congo red staining. Biotech. Histochem. 2018, 93, 543–556. [Google Scholar] [CrossRef]
- Cojocaru, C.; Clima, L. Binding assessment of methylene blue to human serum albumin and poly (acrylic acid): Experimental and computer-aided modeling studies. J. Mol. Liq. 2019, 285, 811–821. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, H.B. Geometry optimization. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2011, 1, 790–809. [Google Scholar] [CrossRef]
- Gazit, E. A possible role for π-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002, 16, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, C.K.; DeForest, C.A. Polymer Design and Development. In Biology and Engineering of Stem Cell Niches; Elsevier: London, UK, 2017; pp. 295–314. [Google Scholar] [CrossRef]
- Choudhuri, S. Chapter 8–Additional Bioinformatic Analyses Involving Protein Sequences. In Bioinformatics for Beginners, 1st ed.; Choudhuri, S., Ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 183–207. [Google Scholar] [CrossRef]
- Cojocaru, C.; Neamtu, A.; Vasiliu, T.; Isac, D.; Pinteala, M. Molecular Dynamics Simulations and In Silico Analysis of Supramolecular Self-Assembled Structures. In New Trends in Macromolecular and Supramolecular Chemistry for Biological Applications, 1st ed.; Abadie, M.J., Pinteala, M., Rotaru, A., Eds.; Springer: Cham, Switzerland, 2021; pp. 357–371. [Google Scholar] [CrossRef]
- Ita, K. Polyplexes for gene and nucleic acid delivery: Progress and bottlenecks. Eur. J. Pharm. Sci. 2020, 150, 105358. [Google Scholar] [CrossRef]
- Bae, Y.; Lee, S.; Green, E.S.; Park, J.H.; Ko, K.S.; Han, J.; Choi, J.S. Characterization of basic amino acids-conjugated PAMAM dendrimers as gene carriers for human adipose-derived mesenchymal stem cells. Int. J. Pharm. 2016, 501, 75–86. [Google Scholar] [CrossRef]
- Yang, J.; Luo, G.-F. Peptide-Based Vectors for Gene Delivery. Chemistry 2023, 5, 1696–1718. [Google Scholar] [CrossRef]
- Vasiliu, T.; Cojocaru, C.; Rotaru, A.; Pricope, G.; Pinteala, M.; Clima, L. Optimization of Polyplex Formation between DNA Oligonucleotide and Poly(ʟ-Lysine): Experimental Study and Modeling Approach. Int. J. Mol. Sci. 2017, 18, 1291. [Google Scholar] [CrossRef]
- Krchňák, V.; Vágner, J.; Šafář, P.; Lebl, M. Noninvasive continuous monitoring of solid-phase peptide synthesis by acid-base indicator. Collect. Czechoslov. Chem. Commun. 1988, 53, 2542–2548. [Google Scholar] [CrossRef]
- Niyangoda, C.; Miti, T.; Breydo, L.; Uversky, V.; Muschol, M. Carbonyl-based blue autofluorescence of proteins and amino acids. PLoS ONE 2017, 12, e0176983. [Google Scholar] [CrossRef]











| FEYNF Peptide Chain | Torsion Angles 1 | |||||
|---|---|---|---|---|---|---|
| Φ1 | Ψ1 | Φ2 | Ψ2 | Φ3 | Ψ3 | |
| Chain-1 | −137.069° | 144.980° | −137.509° | 123.895° | −126.403° | 151.016° |
| Chain-2 | −81.123° | 132.445° | −139.163° | 116.446° | −43.919° | −52.269° |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jitaru, S.-C.; Enache, A.-C.; Cojocaru, C.; Drochioiu, G.; Petre, B.-A.; Gradinaru, V.-R. Self-Assembly of a Novel Pentapeptide into Hydrogelated Dendritic Architecture: Synthesis, Properties, Molecular Docking and Prospective Applications. Gels 2024, 10, 86. https://doi.org/10.3390/gels10020086
Jitaru S-C, Enache A-C, Cojocaru C, Drochioiu G, Petre B-A, Gradinaru V-R. Self-Assembly of a Novel Pentapeptide into Hydrogelated Dendritic Architecture: Synthesis, Properties, Molecular Docking and Prospective Applications. Gels. 2024; 10(2):86. https://doi.org/10.3390/gels10020086
Chicago/Turabian StyleJitaru, Stefania-Claudia, Andra-Cristina Enache, Corneliu Cojocaru, Gabi Drochioiu, Brindusa-Alina Petre, and Vasile-Robert Gradinaru. 2024. "Self-Assembly of a Novel Pentapeptide into Hydrogelated Dendritic Architecture: Synthesis, Properties, Molecular Docking and Prospective Applications" Gels 10, no. 2: 86. https://doi.org/10.3390/gels10020086