Defining Functions of Mannoproteins in Saccharomyces cerevisiae by High-Dimensional Morphological Phenotyping
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Fluorescence Staining, Microscopy, and Image Processing
2.3. Data Analysis
2.4. Dimensionality Reduction and Clustering
2.5. Kinetics of Cluster V Mannoproteins (Dse2 and Egt2)
2.5.1. Yeast Media and Culture Conditions
2.5.2. Constructions of Strains
2.5.3. Primers and Plasmids
2.5.4. Imaging and Data Analysis
2.6. Biochemistry
2.7. Similarity of Mannoprotein Mutants and Drug-Treated Wild-Type Cells in Morphology
2.8. Mannoprotein Analysis Based on Omics Studies
2.8.1. Estimation of Fitness
2.8.2. Analysis of Protein Abundance and Protein–Protein Interactions
2.8.3. Genetic Interaction Analysis
2.8.4. Chemical-Genetic Profile Analysis
3. Results
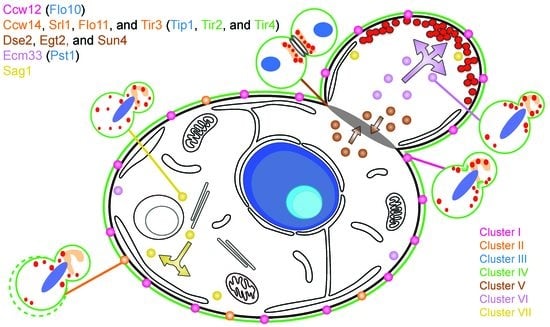
3.1. Effects of Genetic Perturbations on Cell Morphology
3.2. Phenotype of the Cluster I Mutant (ccw12Δ)
3.3. Phenotype of Cluster II Mutants (ccw14Δ, srl1Δ, flo11Δ, and tir3Δ)
3.4. Phenotype of Cluster V Mutants (dse2∆, egt2∆, and sun4∆)
3.5. Phenotype of the Cluster VI Mutant (ecm33∆)
3.6. Phenotype of the Cluster VII Mutant (sag1∆)
3.7. Mannoprotein Gene Duplication
3.8. Comparisons of Morphology and Fitness among Mannoprotein Mutants
3.9. Comparisons of Mannoprotein Mutants and Glycosylation-Defective Cells in Morphology
3.10. Comparison of Morphological Clustering Results with Those from Analyses of Other Omics Data
4. Discussion
4.1. Ccw12 Is a Major Cell Wall Stabilizer
4.2. Cluster V Member Genes Encode Endoglucanases
4.3. ECM33 Plays a Role in Bud Growth
4.4. SAG1 Deletion Perturbs Actin Distribution during Vegetative Growth
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klis, F.M.; Boorsma, A.; De Groot, P.W.J. Cell Wall Construction in Saccharomyces cerevisiae. Yeast 2006, 23, 185–202. [Google Scholar] [CrossRef] [PubMed]
- Lesage, G.; Bussey, H. Cell Wall Assembly in Saccharomyces cerevisiae. Am. Soc. Microbiol. 2006, 70, 317–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orlean, P. Architecture and Biosynthesis of the Saccharomyces cerevisiae Cell Wall. Genetics 2012, 192, 775–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, C.M.; Foor, F.; Marrinan, J.A.; Morin, N.; Nielsen, J.B.; Dahl, A.M.; Mazur, P.; Baginsky, W.; Li, W.; El-Sherbeini, M. The Saccharomyces cerevisiae FKS1 (ETG1) Gene Encodes an Integral Membrane Protein Which Is a Subunit of 1, 3-Beta-D-Glucan Synthase. Proc. Natl. Acad. Sci. USA 1994, 91, 12907–12911. [Google Scholar] [CrossRef] [Green Version]
- Inoue, S.B.; Takewakt, N.; Takasuka, T.; Mio, T.; Adachi, M.; Fujii, Y.; Miyamoto, C.; Arisawa, M.; Furuichi, Y.; Watanabe, T. Characterization and Gene Cloning of 1, 3-β-d-Glucan Synthase from Saccharomyces cerevisiae. Eur. J. Biochem. 1995, 231, 845–854. [Google Scholar] [CrossRef]
- Levin, D.E. Regulation of Cell Wall Biogenesis in Saccharomyces cerevisiae: The Cell Wall Integrity Signaling Pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Groot, P.W.J.; Ruiz, C.; de Aldana, C.R.; Duevnas, E.; Cid, V.J.; Del Rey, F.; Rodriquez-Pena, J.M.; Pérez, P.; Andel, A.; Caubin, J.; et al. A Genomic Approach for the Identification and Classification of Genes Involved in Cell Wall Formation and Its Regulation in Saccharomyces cerevisiae. Comp. Funct. Genom. 2001, 2, 124–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, M.; Goddard, N.; Hicks, C.; Ovalle, R.; Rauceo, J.M.; Jue, C.K.; Lipke, P.N. A Screen for Deficiencies in GPI-Anchorage of Wall Glycoproteins in Yeast. Yeast 2010, 27, 583–596. [Google Scholar] [CrossRef] [Green Version]
- Levy, S.F.; Siegal, M.L. Network Hubs Buffer Environmental Variation in Saccharomyces cerevisiae. PLoS Biol. 2008, 6, e264. [Google Scholar] [CrossRef] [Green Version]
- Warringer, J.; Ericson, E.; Fernandez, L.; Nerman, O.; Blomberg, A. High-Resolution Yeast Phenomics Resolves Different Physiological Features in the Saline Response. Proc. Natl. Acad. Sci. USA 2003, 100, 15724–15729. [Google Scholar] [CrossRef] [Green Version]
- Diss, G.; Gagnon-Arsenault, I.; Dion-Coté, A.M.; Vignaud, H.; Ascencio, D.I.; Berger, C.M.; Landry, C.R. Gene Duplication Can Impart Fragility, Not Robustness, in the Yeast Protein Interaction Network. Science 2017, 355, 630–634. [Google Scholar] [CrossRef]
- Suzuki, G.; Wang, Y.; Kubo, K.; Hirata, E.; Ohnuki, S.; Ohya, Y. Global Study of Holistic Morphological Effectors in the Budding Yeast Saccharomyces cerevisiae. BMC Genom. 2018, 19, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Sipiczki, M.; Strahl, S. Characterization of Ccw12p, a Major Key Player in Cell Wall Stability of Saccharomyces cerevisiae. Yeast 2007, 24, 309–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohya, Y.; Sese, J.; Yukawa, M.; Sano, F.; Nakatani, Y.; Saito, T.L.; Saka, A.; Fukuda, T.; Ishihara, S.; Oka, S.; et al. High-Dimensional and Large-Scale Phenotyping of Yeast Mutants. Proc. Natl. Acad. Sci. USA 2005, 102, 19015–19020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghanegolmohammadi, F.; Yoshida, M.; Ohnuki, S.; Sukegawa, Y.; Okada, H.; Obara, K.; Kihara, A.; Suzuki, K.; Kojima, T.; Yachie, N.; et al. Systematic Analysis of Ca2+ Homeostasis in Saccharomyces cerevisiae Based on Chemical-Genetic Interaction Profiles. Mol. Biol. Cell 2017, 28, 3415–3427. [Google Scholar] [CrossRef]
- Ohnuki, S.; Kashima, M.; Yamada, T.; Ghanegolmohammadi, F.; Zhou, Y.; Goshima, T.; Maruyama, J.-I.; Kitamoto, K.; Hirata, D.; Akao, T.; et al. Genome Editing to Generate Nonfoam-Forming Sake Yeast Strains. Biosci. Biotechnol. Biochem. 2019, 83, 1583–1593. [Google Scholar] [CrossRef] [PubMed]
- Ohnuki, S.; Ohya, Y. High-Dimensional Single-Cell Phenotyping Reveals Extensive Haploinsufficiency. PLoS Biol. 2018, 16, e2005130. [Google Scholar] [CrossRef] [Green Version]
- Chadani, T.; Ohnuki, S.; Isogai, A.; Goshima, T.; Kashima, M.; Ghanegolmohammadi, F.; Nishi, T.; Hirata, D.; Watanabe, D.; Kitamoto, K. Genome Editing to Generate Sake Yeast Strains with Eight Mutations That Confer Excellent Brewing Characteristics. Cells 2021, 10, 1299. [Google Scholar] [CrossRef]
- Nelder, J.A.; Wedderburn, R.W.M. Generalized Linear Models. J. R. Stat. Soc. Ser. A 1972, 135, 370–384. [Google Scholar] [CrossRef]
- Rigby, R.A.; Stasinopoulos, M.D.; Heller, G.Z.; De Bastiani, F. Distributions for Modeling Location, Scale, and Shape: Using GAMLSS in R; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Caicedo, J.C.; Cooper, S.; Heigwer, F.; Warchal, S.; Qiu, P.; Molnar, C.; Vasilevich, A.S.; Barry, J.D.; Bansal, H.S.; Kraus, O.; et al. Data-Analysis Strategies for Image-Based Cell Profiling. Nat. Methods 2017, 14, 849–863. [Google Scholar] [CrossRef]
- Ghanegolmohammadi, F.; Ohnuki, S.; Ohya, Y. Single-Cell Phenomics in Budding Yeast: Technologies and Applications. In Single-Cell Omics; Academic Press: Cambridge, MA, USA, 2019; pp. 355–379. [Google Scholar]
- Scrucca, L.; Fop, M.; Murphy, T.B.; Raftery, A.E. McLust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J. 2016, 8, 289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fink, G.R.; Guthrie, C. Guide to Yeast Genetics and Molecular Biology; Academic Press: Boca Raton, FL, USA, 1991. [Google Scholar]
- Longtine, M.S.; Mckenzie, A., III; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional Modules for Versatile and Economical PCR-Based Gene Deletion and Modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Lee, S.; Lim, W.A.; Thorn, K.S. Improved Blue, Green, and Red Fluorescent Protein Tagging Vectors for S. cerevisiae. PLoS ONE 2013, 8, e67902. [Google Scholar] [CrossRef] [PubMed]
- Slubowski, C.J.; Funk, A.D.; Roesner, J.M.; Paulissen, S.M.; Huang, L.S. Plasmids for C-Terminal Tagging in Saccharomyces cerevisiae That Contain Improved GFP Proteins, Envy and Ivy. Yeast 2015, 32, 379–387. [Google Scholar] [CrossRef] [Green Version]
- Okada, H.; MacTaggart, B.; Ohya, Y.; Bi, E. The Kinetic Landscape and Interplay of Protein Networks in Cytokinesis. Iscience 2021, 24, 101917. [Google Scholar] [CrossRef]
- Markus, S.M.; Omer, S.; Baranowski, K.; Lee, W.L. Improved Plasmids for Fluorescent Protein Tagging of Microtubules in Saccharomyces cerevisiae. Traffic 2015, 16, 773–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onishi, M.; Ko, N.; Nishihama, R.; Pringle, J.R. Distinct Roles of Rho1, Cdc42, and Cyk3 in Septum Formation and Abscission During Yeast Cytokinesis. J. Cell Biol. 2013, 202, 311–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, H.; MacTaggart, B.; Bi, E. Analysis of Local Protein Accumulation Kinetics by Live-Cell Imaging in Yeast Systems. STAR Protoc. 2021, 2, 100733. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Jonasson, E.M.; Rossio, V.; Hatakeyama, R.; Abe, M.; Ohya, Y.; Yoshida, S. Zds1/Zds2–PP2ACdc55 Complex Specifies Signaling Output from Rho1 GTPase. J. Cell Biol. 2016, 212, 51–61. [Google Scholar] [CrossRef] [Green Version]
- Okada, H.; Ohnuki, S.; Roncero, C.; Konopka, J.B.; Ohya, Y. Distinct Roles of Cell Wall Biogenesis in Yeast Morphogenesis As Revealed by Multivariate Analysis of High-Dimensional Morphometric Data. Mol. Biol. Cell 2014, 25, 222–233. [Google Scholar] [CrossRef] [Green Version]
- Storey, J.D.; Bass, A.J.; Dabney, A.; Robinson, D. Qvalue: Q-Value Estimation for False Discovery Rate Control. R Package Version 2.24.0. 2021. Available online: http://github.com/jdstorey/qvalue (accessed on 13 September 2021).
- Ho, B.; Baryshnikova, A.; Brown, G.W. Unification of Protein Abundance Datasets Yields a Quantitative Saccharomyces cerevisiae Proteome. Cell Syst. 2018, 6, 192–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oughtred, R.; Stark, C.; Breitkreutz, B.-J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID Interaction Database: 2019 Update. Nucleic Acids Res. 2019, 47, 529–541. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Costanzo, M.; VanderSluis, B.; Koch, E.N.; Baryshnikova, A.; Pons, C.; Tan, G.; Wang, W.; Usaj, M.; Hanchard, J.; Lee, S.D.; et al. A Global Genetic Interaction Network Maps a Wiring Diagram of Cellular Function. Science 2016, 353, aa1420. [Google Scholar] [CrossRef] [PubMed]
- González-Rubio, G.; Fernández-Acero, T.; Martín, H.; Molina, M. Mitogen-Activated Protein Kinase Phosphatases (MKPs) in Fungal Signaling: Conservation, Function, and Regulation. Int. J. Mol. Sci. 2019, 20, 1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-Gutiérrez, E.; Alegría-Carrasco, E.; Alonso-Rodríguez, E.; Fernández-Acero, T.; Molina, M.; Martín, H. Rewiring the Yeast Cell Wall Integrity (CWI) Pathway through a Synthetic Positive Feedback Circuit Unveils a Novel Role for the MAPKKK Ssk2 in CWI Pathway Activation. FEBS J. 2020, 287, 4881–4901. [Google Scholar] [CrossRef]
- González-Rubio, G.; Sellers-Moya, Á.; Martín, H.; Molina, M. Differential Role of Threonine and Tyrosine Phosphorylation in the Activation and Activity of the Yeast MAPK Slt2. Int. J. Mol. Sci. 2021, 22, 1110. [Google Scholar] [CrossRef]
- Doi, S.; Tanabe, K.; Watanabe, M.; Yamaguchi, M.; Yoshimura, M. An α-Specific Gene, SAG1 Is Required for Sexual Agglutination in Saccharomyces cerevisiae. Curr. Genet. 1989, 15, 393–398. [Google Scholar] [CrossRef]
- Huang, G.; Dougherty, S.D.; Erdman, S.E. Conserved WCPL and CX4C Domains Mediate Several Mating Adhesin Interactions in Saccharomyces cerevisiae. Genetics 2009, 182, 173–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubo, K.; Okada, H.; Shimamoto, T.; Kimori, Y.; Mizunuma, M.; Bi, E.; Ohnuki, S.; Ohya, Y. Implications of Maintenance of Mother–Bud Neck Size in Diverse Vital Processes of Saccharomyces cerevisiae. Curr. Genet. 2019, 65, 253–267. [Google Scholar] [CrossRef]
- Ohnuki, S.; Kobayashi, T.; Ogawa, H.; Kozone, I.; Ueda, J.-Y.; Takagi, M.; Shin, K.-Y.; Hirata, D.; Nogami, S.; Ohya, Y. Analysis of the Biological Activity of a Novel 24-Membered Macrolide JBIR-19 in Saccharomyces cerevisiae by the Morphological Imaging Program CalMorph. FEMS Yeast Res. 2012, 12, 293–304. [Google Scholar] [CrossRef] [Green Version]
- Ragni, E.; Piberger, H.; Neupert, C.; Garcia-Cantalejo, J.; Popolo, L.; Arroyo, J.; Aebi, M.; Strahl, S. The Genetic Interaction Network of CCW12, a Saccharomyces cerevisiae Gene Required for Cell Wall Integrity during Budding and Formation of Mating Projections. BMC Genom. 2011, 12, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Juanes, M.A.; Piatti, S. The Final Cut: Cell Polarity Meets Cytokinesis at the Bud Neck in S. cerevisiae. Cell. Mol. Life Sci. 2016, 73, 3115–3136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, E.L. Mitotic Exit and Separation of Mother and Daughter Cells. Genetics 2012, 192, 1165–1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colman-Lerner, A.; Chin, T.E.; Brent, R. Yeast Cbk1 and Mob2 Activate Daughter-Specific Genetic Programs to Induce Asymmetric Cell Fates. Cell 2001, 107, 739–750. [Google Scholar] [CrossRef] [Green Version]
- Kuznetsov, E.; Váchová, L.; Palková, Z. Cellular Localization of Sun4p and Its Interaction with Proteins in the Yeast Birth Scar. Cell Cycle 2016, 15, 1898–1907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Bernard, A.; Orlando, D.A.; Haase, S.B.; Hartemink, A.J. Branching Process Deconvolution Algorithm Reveals a Detailed Cell-Cycle Transcription Program. Proc. Natl. Acad. Sci. USA 2013, 110, 968–977. [Google Scholar] [CrossRef] [Green Version]
- Lussier, M.; White, A.-M.; Sheraton, J.; di Paolo, T.; Treadwell, J.; Southard, S.B.; Horenstein, C.I.; Chen-Weiner, J.; Ram, A.F.J.; Kapteyn, J.C.; et al. Large Scale Identification of Genes Involved in Cell Surface Biosynthesis and Architecture in Saccharomyces cerevisiae. Genetics 1997, 147, 435–450. [Google Scholar] [CrossRef]
- Pardo, M.; Monteoliva, L.; Vazquez, P.; Martinez, R.; Molero, G.; Nombela, C.; Gil, C. PST1 and ECM33 Encode Two Yeast Cell Surface GPI Proteins Important for Cell Wall Integrity. Microbiology 2004, 150, 4157–4170. [Google Scholar] [CrossRef] [Green Version]
- Bidlingmaier, S.; Snyder, M. Large-Scale Identification of Genes Important for Apical Growth in Saccharomyces cerevisiae by Directed Allele Replacement Technology (DART) Screening. Funct. Integr. Genom. 2002, 1, 345–356. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghanegolmohammadi, F.; Okada, H.; Liu, Y.; Itto-Nakama, K.; Ohnuki, S.; Savchenko, A.; Bi, E.; Yoshida, S.; Ohya, Y. Defining Functions of Mannoproteins in Saccharomyces cerevisiae by High-Dimensional Morphological Phenotyping. J. Fungi 2021, 7, 769. https://doi.org/10.3390/jof7090769
Ghanegolmohammadi F, Okada H, Liu Y, Itto-Nakama K, Ohnuki S, Savchenko A, Bi E, Yoshida S, Ohya Y. Defining Functions of Mannoproteins in Saccharomyces cerevisiae by High-Dimensional Morphological Phenotyping. Journal of Fungi. 2021; 7(9):769. https://doi.org/10.3390/jof7090769
Chicago/Turabian StyleGhanegolmohammadi, Farzan, Hiroki Okada, Yaxuan Liu, Kaori Itto-Nakama, Shinsuke Ohnuki, Anna Savchenko, Erfei Bi, Satoshi Yoshida, and Yoshikazu Ohya. 2021. "Defining Functions of Mannoproteins in Saccharomyces cerevisiae by High-Dimensional Morphological Phenotyping" Journal of Fungi 7, no. 9: 769. https://doi.org/10.3390/jof7090769